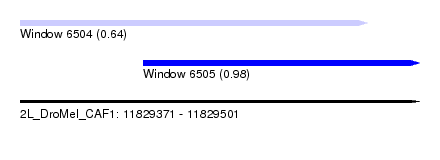
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,829,371 – 11,829,501 |
| Length | 130 |
| Max. P | 0.977388 |

| Location | 11,829,371 – 11,829,484 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -27.05 |
| Energy contribution | -28.58 |
| Covariance contribution | 1.53 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
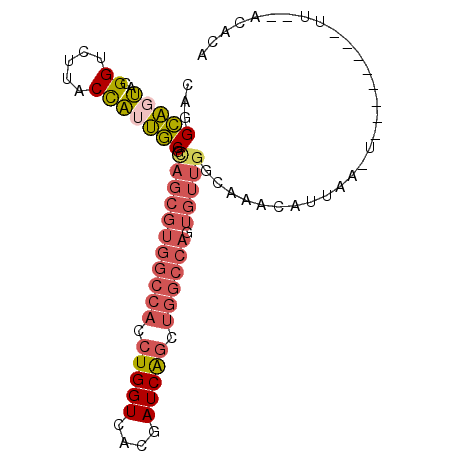
>2L_DroMel_CAF1 11829371 113 + 22407834 GUGACCCCAGUCACGGAUCGCGUAAAAUACCCAGGCAUUG--CAGGCAGUACGGUCUUACCAUUGCGCAGCGUGGCCACCUGGUCA-----CGAUCGGCUGGCCAGUGUUGGUAAACAUU (((((....))))).((((((((((.........(.((((--....)))).)((.....)).))))))...((((((....)))))-----)))))((....))((((((....)))))) ( -37.00) >DroPse_CAF1 20252 102 + 1 GUGGCCGCAGCUGGAGAUCGCGA-AAAUACUCAGCUAUCGAUCAGGGCGUGUGGUCACACUUUUCGAUAG-GCGGUCGACCACACACAAAUCGAUCGGCGCGC----G------------ (((((((.((((((..((.....-..))..))))))((((((....(.((((((((((.(((......))-)..)).)))))))).)..)))))))))).)))----.------------ ( -36.10) >DroSec_CAF1 52572 113 + 1 GUGACCCCAGUCACAGAUCGCGAAAAAUACCCAGACAUUG--CAGGCAGUACGGUCCUACCAUUGCGCAGCGUGGCCACCUGGUCA-----CGAUCAGCUGGCCAGUGUUGGCAAACAUU (((((....))))).......................(((--(..(((((..((.....))))))).(((((((((((.(((((..-----..))))).)))))).)))))))))..... ( -37.10) >DroSim_CAF1 18139 113 + 1 GUGACCCCAGUCACAGAUCGCGAAAAAUACCCAGGCAUUA--CAGGCAGUACGGUCUUACCAUUGCGCAGCGUGGCCACCUGGUCA-----CGAUCAGCUGGCCAGUGUUGGCAAACAUU (((((....)))))((((((.......(((....((....--...)).))))))))).....((((.(((((((((((.(((((..-----..))))).)))))).)))))))))..... ( -37.31) >DroEre_CAF1 18379 113 + 1 GUGACCCCAGUCACGGAUCGCGUAAAAUAUCGAGACAUUG--CGGGCAGUACGGUCUUACCAUUGCGUAGCGUGGCCACCUGGUCA-----CGAUCAGCUGGCCAGUGUUGGCAAAAAUU (((((....))))).((((((((((......(((((..((--(.....)))..)))))....))))))...((((((....)))))-----))))).((..((....))..))....... ( -38.60) >DroYak_CAF1 19372 113 + 1 GUGACCCCAGUCACAGAUCGCGUAAAAUGCCAAGGCAUUG--CAGGCAGUACGGUCUUACCAGUGCGUAGCGUGGCCACCUGGUCA-----CGAUCAGUUGGCCAGUGUUGGCAAACGUU (((((....))))).....((((....((((((.((...(--(..((.((((((.....)).)))))).)).((((((.(((((..-----..))))).)))))))).)))))).)))). ( -44.30) >consensus GUGACCCCAGUCACAGAUCGCGAAAAAUACCCAGGCAUUG__CAGGCAGUACGGUCUUACCAUUGCGCAGCGUGGCCACCUGGUCA_____CGAUCAGCUGGCCAGUGUUGGCAAACAUU (((((....))))).(((((...............................)))))......((((.(((((((((((.((((((.......)))))).)))))).)))))))))..... (-27.05 = -28.58 + 1.53)



| Location | 11,829,411 – 11,829,501 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 72.62 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -19.77 |
| Energy contribution | -20.83 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11829411 90 + 22407834 CAGGCAGUACGGUCUUACCAUUGCGCAGCGUGGCCACCUGGUCACGAUCGGCUGGCCAGUGUUGGUAAACAUUAA-ACGG-UAUUUA--ACACA ..((((((.(((((........((...))((((((....)))))))))))))).))).(((((((...((.....-...)-)..)))--)))). ( -28.80) >DroSec_CAF1 52612 80 + 1 CAGGCAGUACGGUCCUACCAUUGCGCAGCGUGGCCACCUGGUCACGAUCAGCUGGCCAGUGUUGGCAAACAUUAA--------------ACACA ..........((.....)).((((.(((((((((((.(((((....))))).)))))).))))))))).......--------------..... ( -28.10) >DroSim_CAF1 18179 80 + 1 CAGGCAGUACGGUCUUACCAUUGCGCAGCGUGGCCACCUGGUCACGAUCAGCUGGCCAGUGUUGGCAAACAUUAA--------------ACACA .((((......)))).....((((.(((((((((((.(((((....))))).)))))).))))))))).......--------------..... ( -28.50) >DroEre_CAF1 18419 81 + 1 CGGGCAGUACGGUCUUACCAUUGCGUAGCGUGGCCACCUGGUCACGAUCAGCUGGCCAGUGUUGGCAAAAAUUAG-U--------UU----ACA .((((.....((.....)).((((.(((((((((((.(((((....))))).)))))).)))))))))......)-)--------))----... ( -27.10) >DroYak_CAF1 19412 92 + 1 CAGGCAGUACGGUCUUACCAGUGCGUAGCGUGGCCACCUGGUCACGAUCAGUUGGCCAGUGUUGGCAAACGUUAA-UCGG-UAUUUUAUACACA ......(((..(...((((.(....((((((.(((((((((((((.....).))))))).).))))..)))))).-.)))-))..)..)))... ( -28.70) >DroAna_CAF1 28625 73 + 1 ---GGCGGACGGUCUUACUUGGCCACAGUGUAGCCACAUGGUUACGAUCACCAAU--------------CGAUAAGUCGAUUAUUUU----GAA ---((.((.(((((......)))).....((((((....))))))).)).))(((--------------(((....)))))).....----... ( -21.60) >consensus CAGGCAGUACGGUCUUACCAUUGCGCAGCGUGGCCACCUGGUCACGAUCAGCUGGCCAGUGUUGGCAAACAUUAA_U________UU__ACACA ...(((((..((.....))))))).(((((((((((.(((((....))))).)))))).))))).............................. (-19.77 = -20.83 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:06 2006