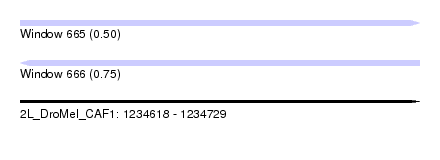
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,234,618 – 1,234,729 |
| Length | 111 |
| Max. P | 0.751047 |

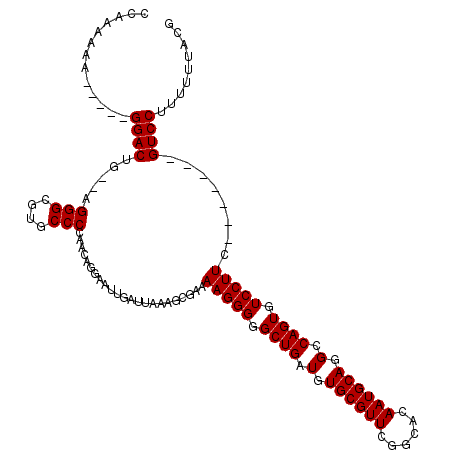
| Location | 1,234,618 – 1,234,729 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -22.77 |
| Energy contribution | -23.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234618 111 + 22407834 CGUAAAAAAGGAC-------GAAGGACACUGGCCUGCAUUGUGCCGAACGCACACCAGCCCCCUUUUCGCUUUAAUCAAUUCCUGUUGGGGCACGCCCU--UAGUCCCUUAGUUUUUUGG ......(((((((-------...((((...(((.(((..(((((.....)))))(((((.........................))))).))).)))..--..))))....))))))).. ( -27.61) >DroSec_CAF1 11940 108 + 1 CGUAAAAAAGGAC-------GAAGGACACUGGCCUGCAUUGUGCCGAACGCACAUCAGCCCCCUUUUCCCUUUAAUCAAUUCCUGUUUGGGCACGCCCUCUCAGUCC-----UUUUUUGC .((((((((((((-------(((((.....((.(((...(((((.....))))).))).)).......)))))..........((...(((....)))...))))))-----)))))))) ( -34.80) >DroSim_CAF1 7757 108 + 1 CGUAAAAAAGGAC-------GAAGGACACUGGCCUGCAUUGUGCCGAACGCACAUCAGCCCCCUUUUCGCUUUAAUCAAUUCCUGUUGGGGCACGCCCUCUCAGUCC-----UUUUUUGC .((((((((((((-------(((((.....((.(((...(((((.....))))).))).))))))).................((..((((....))))..))))))-----)))))))) ( -35.80) >DroYak_CAF1 7985 118 + 1 CGUAAAAAAGGACGAAGGACGAAGGACACUGGCCUGCAUUGUGCCGAACGCACAUCAGCCCCCUUUUCGCUUUAAUCAAUUCCUGUUGGGGCACGCCCU--CAGUCCCUUUUUUUUUUGG ((.((((((((((..(((.((((((.....((.(((...(((((.....))))).))).))))))...((((((((........)))))))).)).)))--..)).))))))))...)). ( -32.50) >consensus CGUAAAAAAGGAC_______GAAGGACACUGGCCUGCAUUGUGCCGAACGCACAUCAGCCCCCUUUUCGCUUUAAUCAAUUCCUGUUGGGGCACGCCCU__CAGUCC_____UUUUUUGC .((((((((((((.......(((((.....((.(((...(((((.....))))).))).)))))))......................(((....))).....)))).....)))))))) (-22.77 = -23.27 + 0.50)

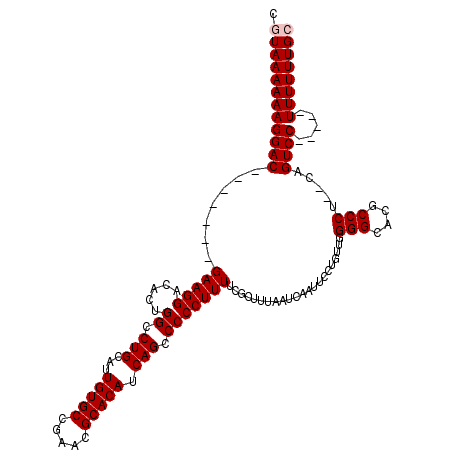
| Location | 1,234,618 – 1,234,729 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.32 |
| Mean single sequence MFE | -38.74 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234618 111 - 22407834 CCAAAAAACUAAGGGACUA--AGGGCGUGCCCCAACAGGAAUUGAUUAAAGCGAAAAGGGGGCUGGUGUGCGUUCGGCACAAUGCAGGCCAGUGUCCUUC-------GUCCUUUUUUACG ..........(((((((.(--(((((...((((........................))))((((((.((((((......)))))).)))))))))))).-------)))))))...... ( -40.46) >DroSec_CAF1 11940 108 - 1 GCAAAAAA-----GGACUGAGAGGGCGUGCCCAAACAGGAAUUGAUUAAAGGGAAAAGGGGGCUGAUGUGCGUUCGGCACAAUGCAGGCCAGUGUCCUUC-------GUCCUUUUUUACG (.((((((-----((((.....(((....))).................(((((.....((.(((.(((((.....)))))...))).))....))))).-------)))))))))).). ( -37.30) >DroSim_CAF1 7757 108 - 1 GCAAAAAA-----GGACUGAGAGGGCGUGCCCCAACAGGAAUUGAUUAAAGCGAAAAGGGGGCUGAUGUGCGUUCGGCACAAUGCAGGCCAGUGUCCUUC-------GUCCUUUUUUACG (.((((((-----((((.((.((((((.(((((........((.....))........)))))...(((((.....)))))...........))))))))-------)))))))))).). ( -35.79) >DroYak_CAF1 7985 118 - 1 CCAAAAAAAAAAGGGACUG--AGGGCGUGCCCCAACAGGAAUUGAUUAAAGCGAAAAGGGGGCUGAUGUGCGUUCGGCACAAUGCAGGCCAGUGUCCUUCGUCCUUCGUCCUUUUUUACG .......((((((((((.(--((((((....((....))................(((((.((((.(.((((((......)))))).).)))).)))))))))))).))))))))))... ( -41.40) >consensus CCAAAAAA_____GGACUG__AGGGCGUGCCCCAACAGGAAUUGAUUAAAGCGAAAAGGGGGCUGAUGUGCGUUCGGCACAAUGCAGGCCAGUGUCCUUC_______GUCCUUUUUUACG .............((((.....(((....))).......................(((((.((((.(.((((((......)))))).).)))).)))))........))))......... (-28.40 = -28.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:31 2006