
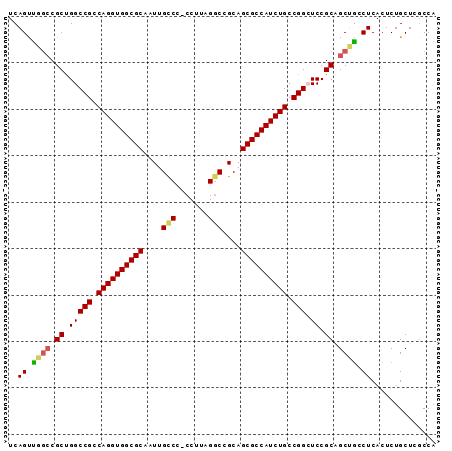
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,816,518 – 11,816,609 |
| Length | 91 |
| Max. P | 0.946215 |

| Location | 11,816,518 – 11,816,609 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -42.12 |
| Consensus MFE | -32.48 |
| Energy contribution | -32.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11816518 91 + 22407834 UCAGUUGGCCGCUGGCCGCCAGGUGGCGCAAUUGUCC-CCUUAGGCCGCAGCGCCAUCUGCCGGCUCCGCAGCUGGCUCACUCUGCUCGCCA ..(((.((((((((((((.((((((((((....(((.-.....)))....)))))))))).))))....)))).)))).))).......... ( -41.60) >DroSec_CAF1 5423 91 + 1 UCAGUUGGCCGCUGGCCGCCAGGUGGCGCUGUUGCCC-CCUUAGUCUGCAGCGCCAUCUGCCGGAUCCGCAGCUACCUCACUCUGCUCGCCA .....((((.((.(((((.(((((((((((((.((..-.....))..))))))))))))).))).)).))(((...........))).)))) ( -39.50) >DroSim_CAF1 5406 90 + 1 UCAGUUGA-CGCUGGCCGCCAGGUGGCGCAGUUGCCC-CCCUAGGCCGCAGCGCCAUCUGCCGGAUCCGCAGCUGCCUCACUCUGCUCGCCA .((((((.-((....(((.((((((((((.((.(((.-.....))).)).)))))))))).)))...))))))))................. ( -39.90) >DroEre_CAF1 5457 90 + 1 UCAGCCUGCCGCCGGCCGACAGGUGGCGCACUGGCC--CCUUAGGCGGCAGCGCCAUCUGCCGGCUCCGCCGCGGGCUCACUCUGCUCUCCA ..(((((((.((.(((((.((((((((((....(((--((...)).))).)))))))))).)))).).)).))))))).............. ( -49.70) >DroYak_CAF1 5528 92 + 1 UCAGCUUGCCGCCGGCCGACAGGUGGCGCCAUGGCCCCCCUUAGGCGGCAGCGCCAUCUGCCGGCUCCGCCGCCGACUCACUCUGCUCGCCA ..(((..((.((.(((((.((((((((((....(((((.....)).))).)))))))))).)))).).)).)).((.....)).)))..... ( -39.90) >consensus UCAGUUGGCCGCUGGCCGCCAGGUGGCGCAAUUGCCC_CCUUAGGCCGCAGCGCCAUCUGCCGGCUCCGCAGCUGCCUCACUCUGCUCGCCA ..((.((((.((.(((((.((((((((((....(((.......)))....)))))))))).))).)).)).)))).)).............. (-32.48 = -32.60 + 0.12)


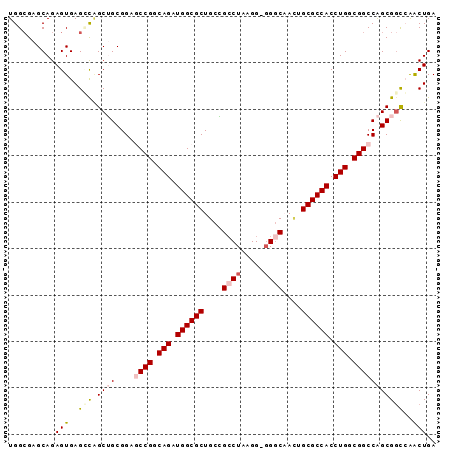
| Location | 11,816,518 – 11,816,609 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 86.87 |
| Mean single sequence MFE | -45.08 |
| Consensus MFE | -32.08 |
| Energy contribution | -33.24 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11816518 91 - 22407834 UGGCGAGCAGAGUGAGCCAGCUGCGGAGCCGGCAGAUGGCGCUGCGGCCUAAGG-GGACAAUUGCGCCACCUGGCGGCCAGCGGCCAACUGA ((((..((...))..))))(((((.(.((((.(((.((((((....(((.....-)).)....)))))).))).))))).)))))....... ( -43.80) >DroSec_CAF1 5423 91 - 1 UGGCGAGCAGAGUGAGGUAGCUGCGGAUCCGGCAGAUGGCGCUGCAGACUAAGG-GGGCAACAGCGCCACCUGGCGGCCAGCGGCCAACUGA ((((..((((..........))))((..(((.(((.((((((((....((....-))....)))))))).))).)))))....))))..... ( -38.60) >DroSim_CAF1 5406 90 - 1 UGGCGAGCAGAGUGAGGCAGCUGCGGAUCCGGCAGAUGGCGCUGCGGCCUAGGG-GGGCAACUGCGCCACCUGGCGGCCAGCG-UCAACUGA (((((.((((..........))))((..(((.(((.((((((.(..((((....-))))..).)))))).))).)))))..))-)))..... ( -41.90) >DroEre_CAF1 5457 90 - 1 UGGAGAGCAGAGUGAGCCCGCGGCGGAGCCGGCAGAUGGCGCUGCCGCCUAAGG--GGCCAGUGCGCCACCUGUCGGCCGGCGGCAGGCUGA ..............((((.((.((.(.((((((((.((((((....(((.....--)))....)))))).))))))))).)).)).)))).. ( -50.20) >DroYak_CAF1 5528 92 - 1 UGGCGAGCAGAGUGAGUCGGCGGCGGAGCCGGCAGAUGGCGCUGCCGCCUAAGGGGGGCCAUGGCGCCACCUGUCGGCCGGCGGCAAGCUGA .......(((..((.(((((((((...)))(((((.((((((((((.((....)).)))...))))))).))))).))))))..))..))). ( -50.90) >consensus UGGCGAGCAGAGUGAGCCAGCUGCGGAGCCGGCAGAUGGCGCUGCCGCCUAAGG_GGGCAACUGCGCCACCUGGCGGCCAGCGGCCAACUGA ..........(((..(((.((((....((((.(((.((((((....((((.....))))....)))))).))).)))))))))))..))).. (-32.08 = -33.24 + 1.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:02 2006