
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,812,112 – 11,812,223 |
| Length | 111 |
| Max. P | 0.583530 |

| Location | 11,812,112 – 11,812,223 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.29 |
| Mean single sequence MFE | -37.75 |
| Consensus MFE | -23.65 |
| Energy contribution | -24.68 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11812112 111 - 22407834 AGGGUGGCGGCGGU------GACAACUAUGAGAACCAGCAGAAUCUGCAGCACAUCUCCGAGCGAGAGGCGCUGGAGGGCGAGGAGGAUCCGUCGGUCGAUCCCACUGCAGCCACCA ..((((((.(((((------(.............((.((((...)))).((...(((((.((((.....)))))))))))..)).(((((........))))))))))).)))))). ( -47.90) >DroVir_CAF1 11264 111 - 1 AGGGCGGCAGUGGU------GAUAACUUUGAGAAUCAACAGAAUCUGCAGCACAUUUCGGAGCGUGAGGCGCUCGAGGGUGAAGAGGAUCCUUCGGUGGAUCCUACAGCAGCCACCA ..((.(((.(((.(------(.....((((........)))).....)).))).(((((.((((.....)))))))))((....(((((((......)))))))...)).))).)). ( -38.80) >DroGri_CAF1 10425 117 - 1 AGGGCGGCAGUGGCAGCAACGAUAACUUUGAGAAUCAACAGAAUCUGCAGCACAUCUCAGAGCGUGAGGCGCUCGAGGGUGAGGAGGAUCCAUCGGUGGAUCCCACAGCAGCUACAA .........(((((.((..(((.....)))(((.((....)).))))).((.(((((..(((((.....)))))..))))).(..(((((((....)))))))..).)).))))).. ( -41.60) >DroWil_CAF1 10357 111 - 1 AGGGCGGAAGUAAU------GAUAAUUUUGAAAAUCAACAGAAUCUGCAACACAUCUCCGAAAGGGAAGCCCUGGAGGGUGAAGAAGAUCCAUCGGUGGAUCCAACAGCGGCUACAA ...((....))...------....((((((........))))))((((..(((.(((((...((((...)))))))))))).....((((((....)))))).....))))...... ( -28.90) >DroMoj_CAF1 9191 108 - 1 AGGGCAACAGCGAU------ACG---GACAACAACCAACAUAAUCUGCAGCACAUCUCGGAGCGUGAGGCGCUCGAGGGUGAGGAGGAUCCCUCAGUGGAUCCAACGGCAGCAACCA .((((....))...------..(---(.......))........((((..(((.(((((.((((.....)))))))))))).(..((((((......))))))..).))))...)). ( -34.20) >DroAna_CAF1 9196 105 - 1 AGAGCGGC------------GAUAAUUUCGAGAACCAGCAGAAUCUGCAACACAUCUCCGAGCGUGAAGCCCUGGAGGGCGAAGAGGAUCCCUCGGUGGAUCCCACGGCGGCCACCA .....(((------------.........((((....((((...))))......))))((..((((..((((....)))).....((((((......)))))))))).))))).... ( -35.10) >consensus AGGGCGGCAGUGGU______GAUAACUUUGAGAACCAACAGAAUCUGCAGCACAUCUCCGAGCGUGAGGCGCUCGAGGGUGAAGAGGAUCCAUCGGUGGAUCCCACAGCAGCCACCA ..(((.((...................................(((....(((.(((((.((((.....)))))))))))).)))((((((......))))))....)).))).... (-23.65 = -24.68 + 1.03)

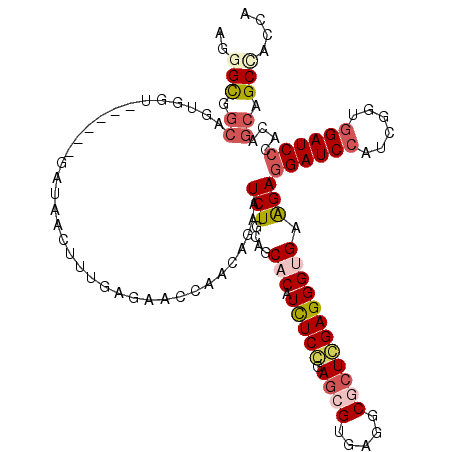
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:59 2006