| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,807,298 – 11,807,391 |
| Length | 93 |
| Max. P | 0.765580 |

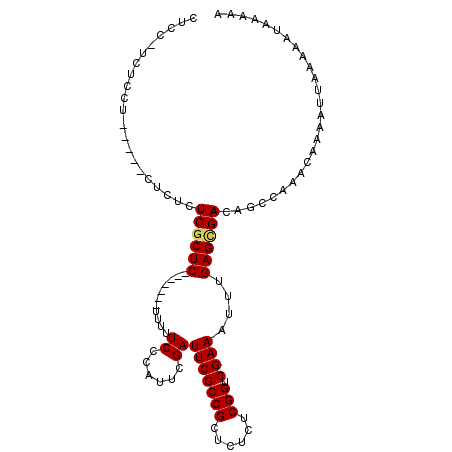
| Location | 11,807,298 – 11,807,391 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -16.77 |
| Energy contribution | -17.15 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11807298 93 + 22407834 UUUUUAUUUUUAAGUUUGUUUGGCUGUCGCUCAAAUUUCGACCGAGAGAGCGGCGAAUCGAAUGGGAAAAA-------GAGUGAGAGAG-----AGGAGAAGGAG (((((((((((...((..(((((.((((((((...((((....))))))))))))..)))))..))...))-------)))))))))..-----........... ( -24.60) >DroSec_CAF1 14614 80 + 1 UUUUUAUUUUUAAUUUUGUUUGGCUGUCGCUCAAAUUUCGACCGAGAGAGCGGCGAAUCGAAUAGGAAAAA-------GAGCGAGA------------------G .(((..(((((..((((((((((.((((((((...((((....))))))))))))..))))))))))..))-------)))..)))------------------. ( -22.10) >DroSim_CAF1 14539 93 + 1 UUUUUAUUUUUAAUUUUGUUUGGCUGUCGCUCAAAUUUCGACCGAGAGAGCGGCGAAUCGAAUAGGAAAAA-------GAGCGAGAGAG-----UAGAGAUGGAG ..((((((((((.((((((((...((((((((...((((....))))))))))))..((......))....-------))))))))...-----)))))))))). ( -24.80) >DroEre_CAF1 13555 105 + 1 UUUUUAUUUUUAAUUUUGUUUGGCUGUCACUCAAAUUUCGACCGAGAGAGCGGCGAAUCGAAUGGGAAAAAGGAAAAAGAGCGAGAGAGAGAGGAGGAGGCGGAG (((((.(((((..((((((((..((....((((...((((.(((......))))))).....)))).....)).....))))))))..))))).)))))...... ( -18.90) >consensus UUUUUAUUUUUAAUUUUGUUUGGCUGUCGCUCAAAUUUCGACCGAGAGAGCGGCGAAUCGAAUAGGAAAAA_______GAGCGAGAGAG_____AGGAGA_GGAG .......................((.((((((....((((.(((......)))))))((......))...........)))))).)).................. (-16.77 = -17.15 + 0.38)

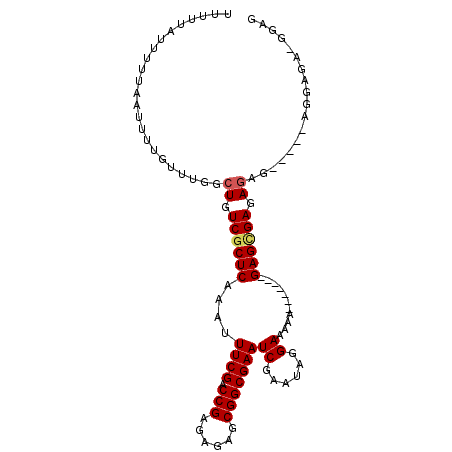
| Location | 11,807,298 – 11,807,391 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -13.48 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.47 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765580 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11807298 93 - 22407834 CUCCUUCUCCU-----CUCUCUCACUC-------UUUUUCCCAUUCGAUUCGCCGCUCUCUCGGUCGAAAUUUGAGCGACAGCCAAACAAACUUAAAAAUAAAAA .........((-----.((.((((...-------....((......))(((((((......))).))))...)))).)).))....................... ( -10.60) >DroSec_CAF1 14614 80 - 1 C------------------UCUCGCUC-------UUUUUCCUAUUCGAUUCGCCGCUCUCUCGGUCGAAAUUUGAGCGACAGCCAAACAAAAUUAAAAAUAAAAA (------------------(.((((((-------....((......))(((((((......))).))))....)))))).))....................... ( -15.00) >DroSim_CAF1 14539 93 - 1 CUCCAUCUCUA-----CUCUCUCGCUC-------UUUUUCCUAUUCGAUUCGCCGCUCUCUCGGUCGAAAUUUGAGCGACAGCCAAACAAAAUUAAAAAUAAAAA ...........-----..((.((((((-------....((......))(((((((......))).))))....)))))).))....................... ( -15.10) >DroEre_CAF1 13555 105 - 1 CUCCGCCUCCUCCUCUCUCUCUCGCUCUUUUUCCUUUUUCCCAUUCGAUUCGCCGCUCUCUCGGUCGAAAUUUGAGUGACAGCCAAACAAAAUUAAAAAUAAAAA ..................((.((((((...........((......))(((((((......))).))))....)))))).))....................... ( -13.20) >consensus CUCC_UCUCCU_____CUCUCUCGCUC_______UUUUUCCCAUUCGAUUCGCCGCUCUCUCGGUCGAAAUUUGAGCGACAGCCAAACAAAAUUAAAAAUAAAAA .....................((((((...........((......))(((((((......))).))))....)))))).......................... (-11.35 = -11.47 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:56 2006