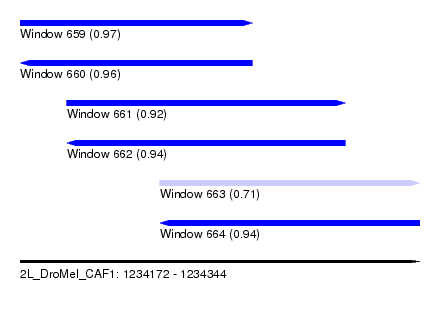
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,234,172 – 1,234,344 |
| Length | 172 |
| Max. P | 0.971631 |

| Location | 1,234,172 – 1,234,272 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -21.27 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234172 100 + 22407834 --------------------CUCCUCUGCCAUCCUUGCCUAAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUUUUUUGGCGAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAA --------------------.((((.((((((....(((..........))).))))))......................((((((.((((.(.....)))))))))))....)))).. ( -25.80) >DroSec_CAF1 11515 95 + 1 -----------------------CUCUGCCAUCCUUGCCUAAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUU--UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAA -----------------------...((((((....(((..........))).))))))........((((.(((....(--(((((((.((.(.....)))))))))))..))))))). ( -25.50) >DroSim_CAF1 7332 95 + 1 -----------------------CUCUGCCAUCCUUGCCUAAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUU--UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAA -----------------------...((((((....(((..........))).))))))........((((.(((....(--(((((((.((.(.....)))))))))))..))))))). ( -25.50) >DroYak_CAF1 7507 118 + 1 CCCAAGUUGUGUGUGCCAUCCUCCGCUGGCAUCCUUGCCUAAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCCUUUCUCUU--UCGGCCAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAA .............((((((...(((..((((....)))).........)))..))))))...........(((((...((--(.(((.((((.(.....)))))))))))..)))))... ( -27.00) >consensus _______________________CUCUGCCAUCCUUGCCUAAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUU__UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAA ..........................((((((....(((..........))).)))))).............((((.(((..(((((.((((.(.....))))))))))....))))))) (-21.27 = -21.77 + 0.50)

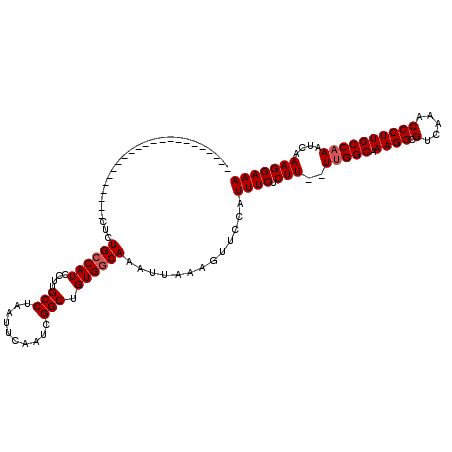
| Location | 1,234,172 – 1,234,272 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.41 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -27.26 |
| Energy contribution | -28.07 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234172 100 - 22407834 UUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUCGCCAAAAAAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUUAGGCAAGGAUGGCAGAGGAG-------------------- ((((((((..((((((.(((((.....))))).)))))))))).)))).....(((...((((((((.(((..........)))...).))))))).)))-------------------- ( -29.30) >DroSec_CAF1 11515 95 - 1 UUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA--AAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUUAGGCAAGGAUGGCAGAG----------------------- .(((((((..((((((((((((.....)))))))))))--)...)))..))))......((((((((.(((..........)))...).))))))).----------------------- ( -32.30) >DroSim_CAF1 7332 95 - 1 UUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA--AAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUUAGGCAAGGAUGGCAGAG----------------------- .(((((((..((((((((((((.....)))))))))))--)...)))..))))......((((((((.(((..........)))...).))))))).----------------------- ( -32.30) >DroYak_CAF1 7507 118 - 1 UUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUGGCCGA--AAGAGAAAGGGAACUUUAAUUUGCCACAGCCGAUUGAAUUAGGCAAGGAUGCCAGCGGAGGAUGGCACACACAACUUGGG ((((((((..((((((.(((((.....))))).)))))--)....))))))))..........(((..(((..........)))..(..(((((........)))))..)......))). ( -33.90) >consensus UUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA__AAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUUAGGCAAGGAUGGCAGAG_______________________ ....((((...(((((((((((.....)))))))))))...))))..............((((((((.(((..........)))...).)))))))........................ (-27.26 = -28.07 + 0.81)

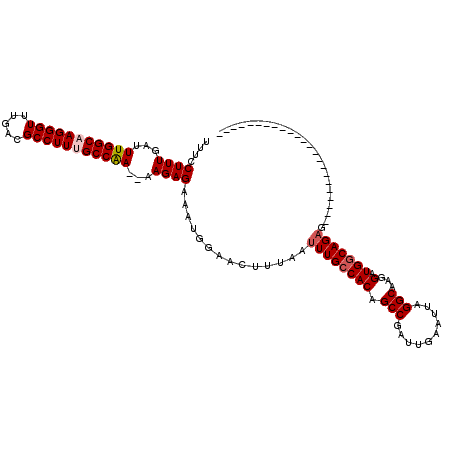
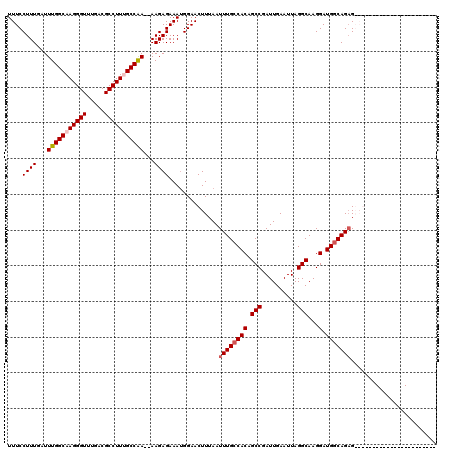
| Location | 1,234,192 – 1,234,312 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -29.65 |
| Energy contribution | -29.96 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234192 120 + 22407834 AAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUUUUUUGGCGAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGUCAAAG .........((((.((((........(((((..........((((((.((((.(.....)))))))))))...(((((....)))))......)))))........).))).)))).... ( -31.89) >DroSec_CAF1 11532 118 + 1 AAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUU--UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGUCAAAG .........((((.((((........(((((........(--(((((((.((.(.....)))))))))))...(((((....)))))......)))))........).))).)))).... ( -31.89) >DroSim_CAF1 7349 118 + 1 AAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUU--UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGCCAAAG .........((((.((((........(((((........(--(((((((.((.(.....)))))))))))...(((((....)))))......)))))........).))).)))).... ( -34.59) >DroYak_CAF1 7547 118 + 1 AAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCCUUUCUCUU--UCGGCCAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGAAAACCCUUGGCCGAGGGUCAAAG .....(((..((....))..)))........((((..(((--((((((((((.((....(((.((......))(((((....)))))......)))...)))))))))))))))..)))) ( -33.80) >consensus AAUUCAAUCGGCUGUGGCAAAUUAAAGUUCCAUUUCUCUU__UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGUCAAAG .........((((.((((........(((((...........(((((.((((.(.....))))))))))....(((((....)))))......)))))........))).).)))).... (-29.65 = -29.96 + 0.31)

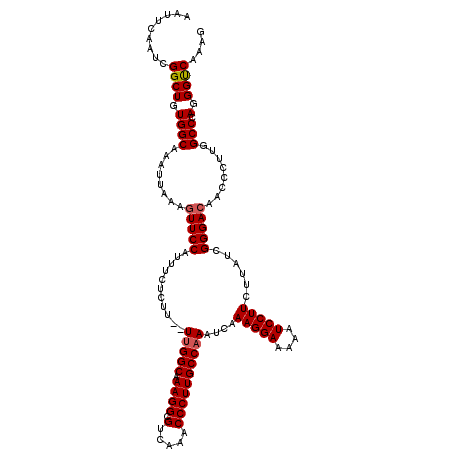
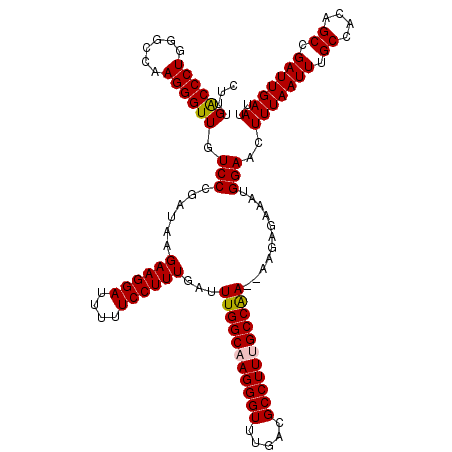
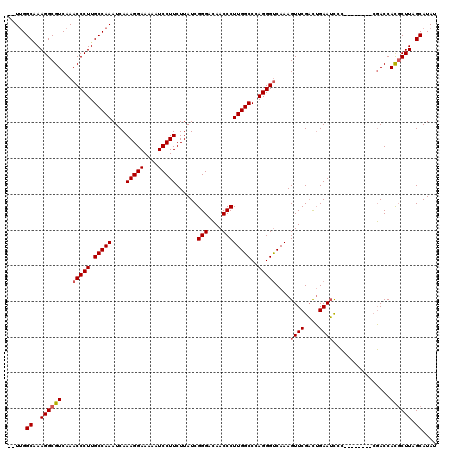
| Location | 1,234,192 – 1,234,312 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.36 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -34.72 |
| Energy contribution | -34.85 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234192 120 - 22407834 CUUUGACCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUCGCCAAAAAAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUU ....((((((......))))))((.(((....((((((....))))))..((((((.(((((.....))))).)))))).........))).))(((((((.((....)).))))))).. ( -35.10) >DroSec_CAF1 11532 118 - 1 CUUUGACCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA--AAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUU ....((((((......))))))((.(((....((((((....))))))..((((((((((((.....)))))))))))--).......))).))(((((((.((....)).))))))).. ( -39.20) >DroSim_CAF1 7349 118 - 1 CUUUGGCCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA--AAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUU ((((((((...))))))))...((.(((....((((((....))))))..((((((((((((.....)))))))))))--).......))).))(((((((.((....)).))))))).. ( -41.90) >DroYak_CAF1 7547 118 - 1 CUUUGACCCUCGGCCAAGGGUUUUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUGGCCGA--AAGAGAAAGGGAACUUUAAUUUGCCACAGCCGAUUGAAUU ......((((((((((((((((..(((..(((((((((....))))))...)))....)))...))).))))))))))--........)))...(((((((.((....)).))))))).. ( -40.30) >consensus CUUUGACCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA__AAGAGAAAUGGAACUUUAAUUUGCCACAGCCGAUUGAAUU ....((((((......)))))).(((......((((((....))))))...(((((((((((.....)))))))))))...........)))..(((((((.((....)).))))))).. (-34.72 = -34.85 + 0.13)

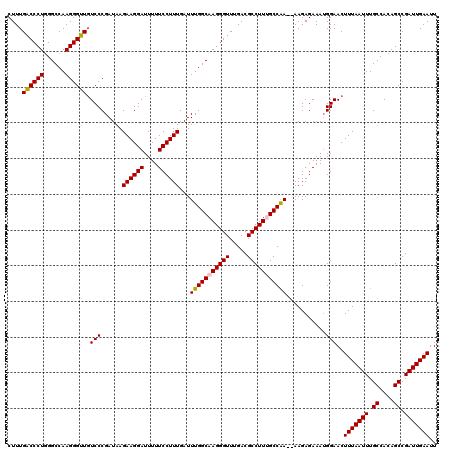
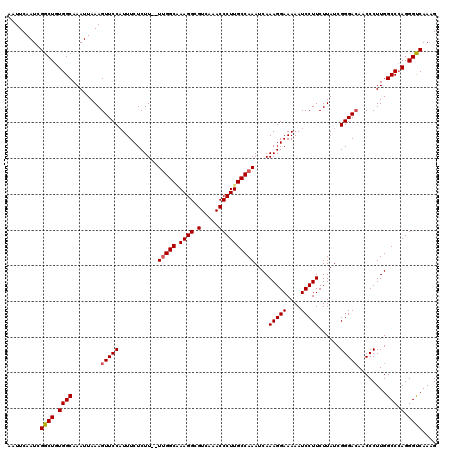
| Location | 1,234,232 – 1,234,344 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.41 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -24.99 |
| Energy contribution | -25.55 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234232 112 + 22407834 UUUUGGCGAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGUCAAAGUUCGACUGAAUCCC--------CGACCACGCUUAGCAUAU .....((..((((((...(((((.(((((....(((((....)))))......(((....))))))))..)))))....(.(((..........--------))).))))))).)).... ( -30.10) >DroSec_CAF1 11572 110 + 1 --UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGUCAAAGUUCGACUGAAUCCC--------CGACCACGCUUAGCAUAU --...((..((((((...(((((.(((((....(((((....)))))......(((....))))))))..)))))....(.(((..........--------))).))))))).)).... ( -28.70) >DroSim_CAF1 7389 110 + 1 --UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGCCAAAGUUCGACUGAAUCCC--------CGACCACGCUUAGCAUAU --(((((((.((.(.....))))))))))....(((((....)))))....(((((.......((((((...)))))).((((....)))).))--------)))............... ( -30.00) >DroYak_CAF1 7587 118 + 1 --UCGGCCAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGAAAACCCUUGGCCGAGGGUCAAAGUUCGCUGGAAGUGGGAGGGGGGAUACCAUCCUUAGCAUAU --(((((...(((.....(((((((((((....(((((....)))))......(((....))))))).)))))))....))).)))))..((..(((((((....)).))))).)).... ( -37.10) >consensus __UUGGCAAAGGCGUCAAACCCUUGCCAAAUCAAAGGAAAAAUCCUUCUUAUCGGGACAACCCUUGGCCCAGGGUCAAAGUUCGACUGAAUCCC________CGACCACGCUUAGCAUAU .....((..((((((...(((((.(((((....(((((....)))))......(((....))))))))..)))))....((((....))))................)))))).)).... (-24.99 = -25.55 + 0.56)

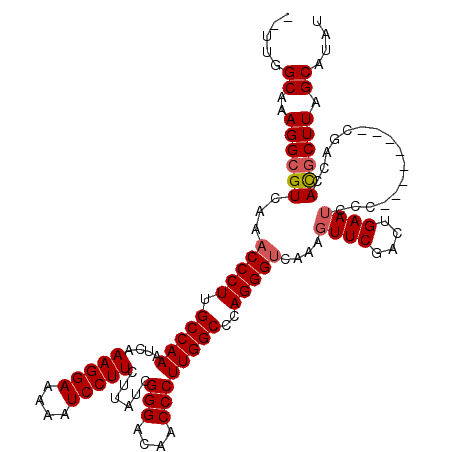
| Location | 1,234,232 – 1,234,344 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.41 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -30.86 |
| Energy contribution | -30.93 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1234232 112 - 22407834 AUAUGCUAAGCGUGGUCG--------GGGAUUCAGUCGAACUUUGACCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUCGCCAAAA .(((((...)))))((((--------(((.(((....)))....((((((......)))))).)))))))..((((((....))))))..((((((.(((((.....))))).)))))). ( -40.10) >DroSec_CAF1 11572 110 - 1 AUAUGCUAAGCGUGGUCG--------GGGAUUCAGUCGAACUUUGACCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA-- .(((((...)))))((((--------(((.(((....)))....((((((......)))))).)))))))..((((((....))))))...(((((((((((.....)))))))))))-- ( -42.40) >DroSim_CAF1 7389 110 - 1 AUAUGCUAAGCGUGGUCG--------GGGAUUCAGUCGAACUUUGGCCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA-- .(((((...)))))((((--------(((.(((....)))((((((((...))))))))....)))))))..((((((....))))))...(((((((((((.....)))))))))))-- ( -45.60) >DroYak_CAF1 7587 118 - 1 AUAUGCUAAGGAUGGUAUCCCCCCUCCCACUUCCAGCGAACUUUGACCCUCGGCCAAGGGUUUUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUGGCCGA-- ...((((..((((((...........)))..)))))))...........(((((((((((((..(((..(((((((((....))))))...)))....)))...))).))))))))))-- ( -38.40) >consensus AUAUGCUAAGCGUGGUCG________GGGAUUCAGUCGAACUUUGACCCUGGGCCAAGGGUUGUCCCGAUAAGAAGGAUUUUUCCUUUGAUUUGGCAAGGGUUUGACGCCUUUGCCAA__ ....((.(((.(((................(((....)))....((((((..((((((((....))).....((((((....))))))...))))).))))))...)))))).))..... (-30.86 = -30.93 + 0.06)

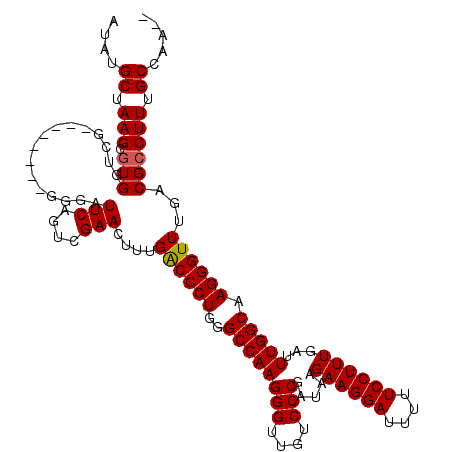
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:29 2006