
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,802,276 – 11,802,502 |
| Length | 226 |
| Max. P | 0.999925 |

| Location | 11,802,276 – 11,802,382 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -29.72 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.12 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802276 106 + 22407834 UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). ( -29.72) >DroSec_CAF1 9630 106 + 1 UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). ( -29.72) >DroSim_CAF1 9623 106 + 1 UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). ( -29.72) >DroWil_CAF1 14675 106 + 1 UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). ( -29.72) >DroYak_CAF1 9535 106 + 1 UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). ( -29.72) >DroPer_CAF1 13835 106 + 1 UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). ( -29.72) >consensus UCACACUCACCAGUGUGCCACAUGUAGUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGC .((.((.(((..(((((((((((((.(((((......((...(((((.......)))))...)).......))))).)).))))))))...)))..))).)).)). (-29.72 = -29.72 + 0.00)



| Location | 11,802,276 – 11,802,382 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802276 106 - 22407834 GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). ( -31.30) >DroSec_CAF1 9630 106 - 1 GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). ( -31.30) >DroSim_CAF1 9623 106 - 1 GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). ( -31.30) >DroWil_CAF1 14675 106 - 1 GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). ( -31.30) >DroYak_CAF1 9535 106 - 1 GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). ( -31.30) >DroPer_CAF1 13835 106 - 1 GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). ( -31.30) >consensus GCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCACUACAUGUGGCACACUGGUGAGUGUGA .....(((((.((((...))))...((.(((((.....((((((......))))))))))).)).((((((((...((((.....)))).))).))))).))))). (-31.30 = -31.30 + 0.00)

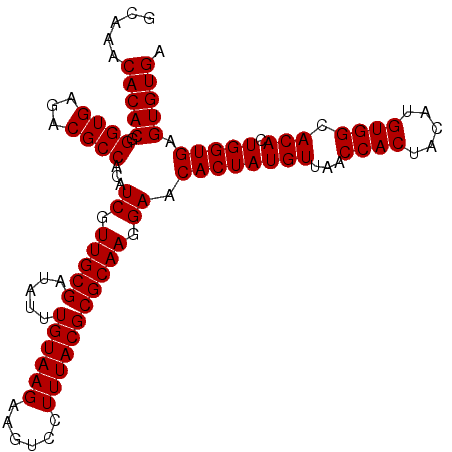

| Location | 11,802,302 – 11,802,422 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -34.95 |
| Energy contribution | -35.12 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802302 120 + 22407834 GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUU (((.....)))(((((((...((((((((.(((...............((((..(..(((.(.....))))..)..))))((((((((....)))))))))))))))))))))))).)). ( -41.60) >DroPse_CAF1 13712 120 + 1 GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUU (((.....)))(((((((...((((((((.(((...............((((..(..(((.(.....))))..)..))))(((.((((....)))).))))))))))))))))))).)). ( -35.60) >DroSim_CAF1 9649 120 + 1 GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUU (((.....)))(((((((...((((((((.(((...............((((..(..(((.(.....))))..)..))))((((((((....)))))))))))))))))))))))).)). ( -41.60) >DroWil_CAF1 14701 120 + 1 GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUAAACGUCUU (((.....)))........((((((((((.(((...............((((..(..(((.(.....))))..)..))))(((.((((....)))).))))))))))))))))....... ( -34.10) >DroYak_CAF1 9561 120 + 1 GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUU (((.....)))(((((((...((((((((.(((...............((((..(..(((.(.....))))..)..))))((((((((....)))))))))))))))))))))))).)). ( -41.60) >DroPer_CAF1 13861 120 + 1 GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUU (((.....)))(((((((...((((((((.(((...............((((..(..(((.(.....))))..)..))))(((.((((....)))).))))))))))))))))))).)). ( -35.60) >consensus GUGGUUAACAUAGUGUUCCUUGCGCGUAAAGGACUUCUUACAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUU (((.....)))(((((((...((((((((.(((...............((((..(..(((.(.....))))..)..))))(((.((((....)))).))))))))))))))))))).)). (-34.95 = -35.12 + 0.17)

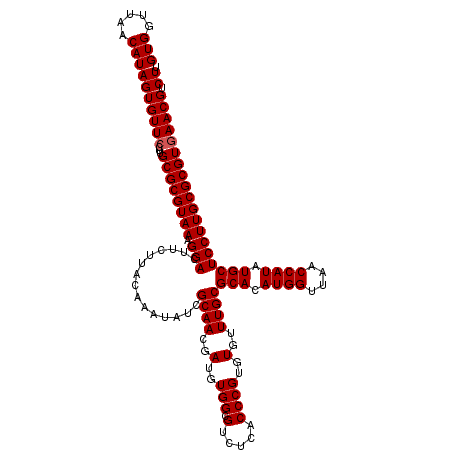
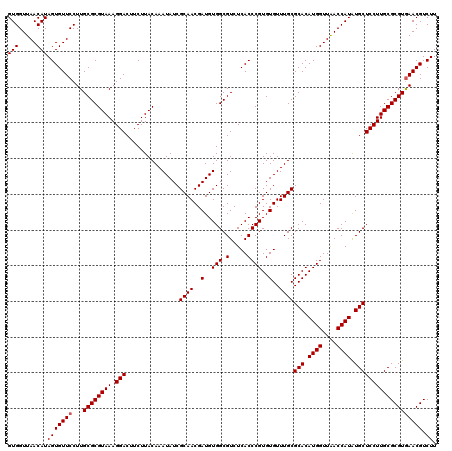
| Location | 11,802,302 – 11,802,422 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -38.16 |
| Energy contribution | -37.77 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802302 120 - 22407834 AAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. ( -39.02) >DroPse_CAF1 13712 120 - 1 AAGACGUUCACGCGCAAGGAGCAUAUGGUUAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. ( -37.12) >DroSim_CAF1 9649 120 - 1 AAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. ( -39.02) >DroWil_CAF1 14701 120 - 1 AAGACGUUUACGCGCAAGGAGCAUAUGGUUAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. ( -35.32) >DroYak_CAF1 9561 120 - 1 AAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. ( -39.02) >DroPer_CAF1 13861 120 - 1 AAGACGUUCACGCGCAAGGAGCAUAUGGUUAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. ( -37.12) >consensus AAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUGUAAGAAGUCCUUUACGCGCAAGGAACACUAUGUUAACCAC .....((((..(((((((((((((((((....))))))))((((.......((((...))))......))))...............)))))...))))...)))).............. (-38.16 = -37.77 + -0.39)

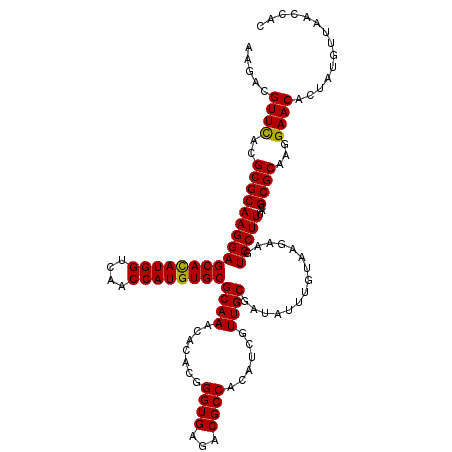

| Location | 11,802,342 – 11,802,462 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -46.28 |
| Consensus MFE | -42.48 |
| Energy contribution | -42.65 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802342 120 + 22407834 CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC (((((((((...))))))(((((.(((.((((((((....)))))))).))).(((((((.....))))))).))))))))(((((...(((....)))..(((((...))))).))))) ( -49.60) >DroPse_CAF1 13752 120 + 1 CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC (((((((((...))))))(((((.((((((((((((....))))))))..........((.....))..))))))))))))(((((...(((....)))..(((((...))))).))))) ( -43.50) >DroSim_CAF1 9689 120 + 1 CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC (((((((((...))))))(((((.(((.((((((((....)))))))).))).(((((((.....))))))).))))))))(((((...(((....)))..(((((...))))).))))) ( -49.60) >DroWil_CAF1 14741 120 + 1 CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUAAACGUCUUUGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC ........((((..(..(((.(.....))))..)..))))((((((((..........((.....))(((.((((((((((.((..((....))..)).))))))))))))))))))))) ( -41.90) >DroYak_CAF1 9601 120 + 1 CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC (((((((((...))))))(((((.(((.((((((((....)))))))).))).(((((((.....))))))).))))))))(((((...(((....)))..(((((...))))).))))) ( -49.60) >DroPer_CAF1 13901 120 + 1 CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC (((((((((...))))))(((((.((((((((((((....))))))))..........((.....))..))))))))))))(((((...(((....)))..(((((...))))).))))) ( -43.50) >consensus CAAAUAUCGCAACGAUGUGGCGUCUCACCCGUGUGUUUGCGCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGC ...((((((...))))))(((((.((((((((((((....))))))))..........((.....))..)))))))))...(((((...(((....)))..(((((...))))).))))) (-42.48 = -42.65 + 0.17)

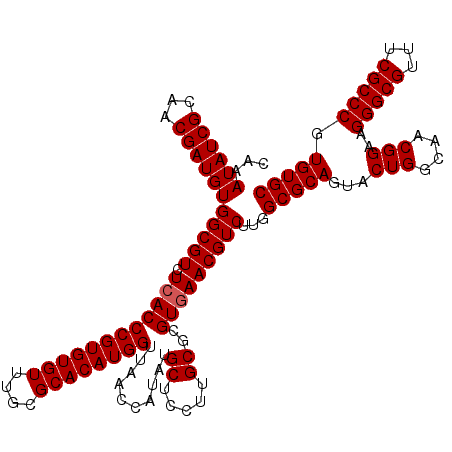
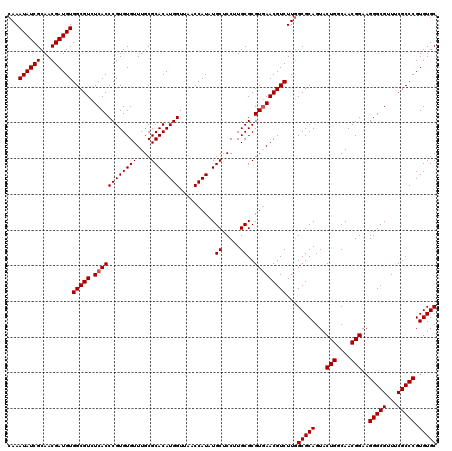
| Location | 11,802,342 – 11,802,462 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.44 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -42.90 |
| Energy contribution | -42.65 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802342 120 - 22407834 GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.....((((......(((((............)))))....((((((((....))))))))))))......)))))...))))....))))))........ ( -43.50) >DroPse_CAF1 13752 120 - 1 GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCAUAUGGUUAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.....((((......(((((............)))))....((((((((....))))))))))))......)))))...))))....))))))........ ( -41.60) >DroSim_CAF1 9689 120 - 1 GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.....((((......(((((............)))))....((((((((....))))))))))))......)))))...))))....))))))........ ( -43.50) >DroWil_CAF1 14741 120 - 1 GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCAAAGACGUUUACGCGCAAGGAGCAUAUGGUUAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.(((.((((..(((..(((......))).)))..)))))))((((((((....))))))))..........)))))...))))....))))))........ ( -43.10) >DroYak_CAF1 9601 120 - 1 GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.....((((......(((((............)))))....((((((((....))))))))))))......)))))...))))....))))))........ ( -43.50) >DroPer_CAF1 13901 120 - 1 GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCAUAUGGUUAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.....((((......(((((............)))))....((((((((....))))))))))))......)))))...))))....))))))........ ( -41.60) >consensus GCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGCGCAAACACACGGGUGAGACGCCACAUCGUUGCGAUAUUUG (((.(((((((...(((((.....((((......(((((............)))))....((((((((....))))))))))))......)))))...))))....))))))........ (-42.90 = -42.65 + -0.25)

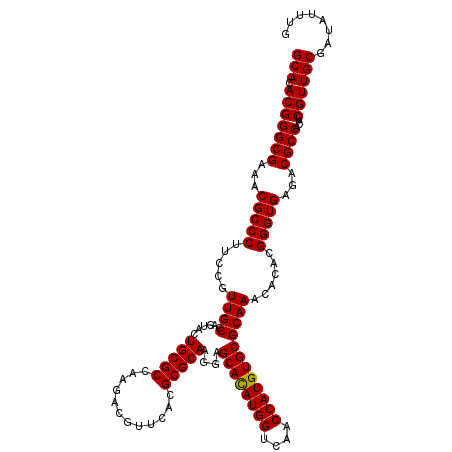
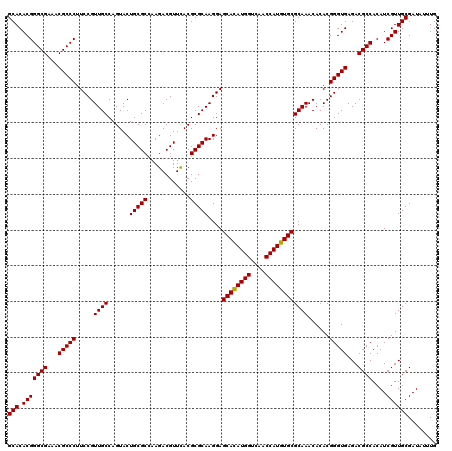
| Location | 11,802,382 – 11,802,502 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -49.78 |
| Consensus MFE | -43.93 |
| Energy contribution | -44.02 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.965007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802382 120 + 22407834 GCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAGCGGAAGUGAUUGUCCAGAUGCUCUUUGCGGGCAAAGG ((((((((....)))))))).((((((.((..(((((((.((((((...(((....)))..(((((...)))))...)))..((....))....))).)))))...))..)).)).)))) ( -53.40) >DroPse_CAF1 13792 120 + 1 GCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAGCGAAAGUGAUUGUCCAGAUGCUCUUUGCGGGCAAAGG (((.((((....)))).))).((((((.((..(((((((.((((((...(((....)))..(((((...)))))...)))..((....))....))).)))))...))..)).)).)))) ( -47.40) >DroSim_CAF1 9729 120 + 1 GCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAGCGGAAGUGAUUGUCCAGAUGCUCUUUGCGGGCAAAGG ((((((((....)))))))).((((((.((..(((((((.((((((...(((....)))..(((((...)))))...)))..((....))....))).)))))...))..)).)).)))) ( -53.40) >DroWil_CAF1 14781 120 + 1 GCACAUGGUUAACCAUAUGCUCCUUGCGCGUAAACGUCUUUGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAACGAAAAUGAUUGUCCAGAUGCUCUUUGCGGGCAAAUG (((.((((....)))).)))...((((.((((((((((((((((((...(((....)))..(((((...))))).)))))))((((......))))..)))))...)))))).))))... ( -44.10) >DroYak_CAF1 9641 120 + 1 GCACAUGGUUGACCAUGUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAGCGGAAGUGAUUGUCCAGGUGCUCUUUGCGGGCAAAGG ((((((((....)))))))).((((((.((..((.(((((((((((...(((....)))..(((((...)))))...)))).((....))......))))).))..))..)).)).)))) ( -53.00) >DroPer_CAF1 13941 120 + 1 GCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAGCGAAAGUGAUUGUCCAGAUGCUCUUUGCGGGCAAAGG (((.((((....)))).))).((((((.((..(((((((.((((((...(((....)))..(((((...)))))...)))..((....))....))).)))))...))..)).)).)))) ( -47.40) >consensus GCACAUGGUUAACCAUAUGCUCCUUGCGCGUGAACGUCUUGGCGCAGUACUGGCAACGGAAGGGCGUUUCGCCCGUGUGCGAGCGAAAGUGAUUGUCCAGAUGCUCUUUGCGGGCAAAGG (((.((((....)))).))).((((((.(((((((((((.((((((...(((....)))..(((((...)))))...)))..((....))....))).)))))...)))))).)).)))) (-43.93 = -44.02 + 0.08)

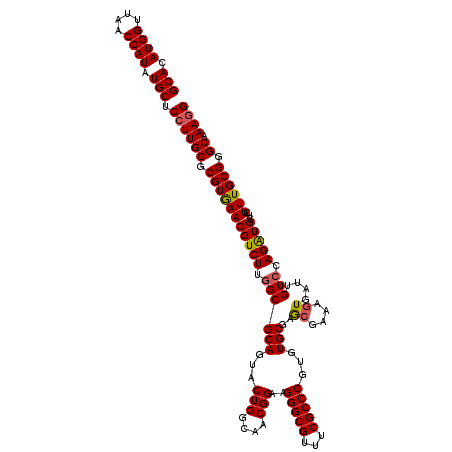
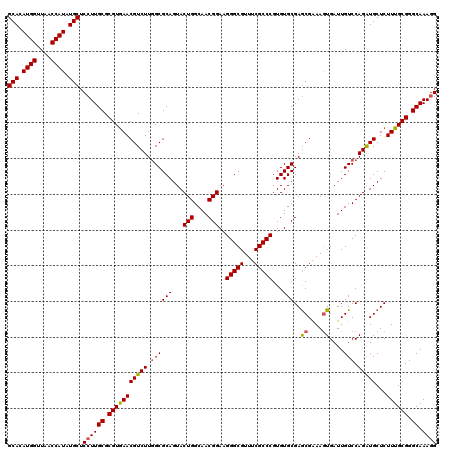
| Location | 11,802,382 – 11,802,502 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -39.07 |
| Energy contribution | -38.43 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11802382 120 - 22407834 CCUUUGCCCGCAAAGAGCAUCUGGACAAUCACUUCCGCUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGC ..((((((((....((((....(((........))))))).....)))))))).......(((.((((..((...(((......)))..))..)))))))((((((((....)))))))) ( -41.20) >DroPse_CAF1 13792 120 - 1 CCUUUGCCCGCAAAGAGCAUCUGGACAAUCACUUUCGCUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCAUAUGGUUAACCAUGUGC .((((((.((....((((.((((((.......(((((((((....)))))))))......)))...((((....)).))..))).)))).)).)))))).((((((((....)))))))) ( -40.42) >DroSim_CAF1 9729 120 - 1 CCUUUGCCCGCAAAGAGCAUCUGGACAAUCACUUCCGCUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGC ..((((((((....((((....(((........))))))).....)))))))).......(((.((((..((...(((......)))..))..)))))))((((((((....)))))))) ( -41.20) >DroWil_CAF1 14781 120 - 1 CAUUUGCCCGCAAAGAGCAUCUGGACAAUCAUUUUCGUUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCAAAGACGUUUACGCGCAAGGAGCAUAUGGUUAACCAUGUGC ..((((((((....((((.....(.....)......)))).....)))))))).......(((.((((..(((..(((......))).)))..)))))))((((((((....)))))))) ( -36.40) >DroYak_CAF1 9641 120 - 1 CCUUUGCCCGCAAAGAGCACCUGGACAAUCACUUCCGCUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGC ..((((((((....((((....(((........))))))).....)))))))).......(((.((((..((...(((......)))..))..)))))))((((((((....)))))))) ( -41.20) >DroPer_CAF1 13941 120 - 1 CCUUUGCCCGCAAAGAGCAUCUGGACAAUCACUUUCGCUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCAUAUGGUUAACCAUGUGC .((((((.((....((((.((((((.......(((((((((....)))))))))......)))...((((....)).))..))).)))).)).)))))).((((((((....)))))))) ( -40.42) >consensus CCUUUGCCCGCAAAGAGCAUCUGGACAAUCACUUCCGCUCGCACACGGGCGAAACGCCCUUCCGUUGCCAGUACUGCGCCAAGACGUUCACGCGCAAGGAGCACAUGGUCAACCAUGUGC ..((((((((....((((....(((........))))))).....)))))))).......(((.((((..((...(((......)))..))..)))))))((((((((....)))))))) (-39.07 = -38.43 + -0.64)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:49 2006