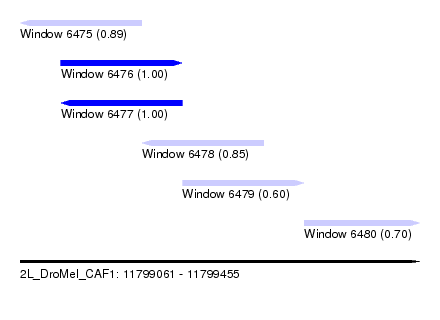
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,799,061 – 11,799,455 |
| Length | 394 |
| Max. P | 0.999975 |

| Location | 11,799,061 – 11,799,181 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -37.12 |
| Consensus MFE | -33.38 |
| Energy contribution | -33.77 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11799061 120 - 22407834 CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAGUCGCCGCACAAGUGCACGUACUGCACCAAGACGUUCACGCGCAAGGAGCACCUGACGAACCAUGUGCGCCAGCACACGGGCGAC (((((((((..(((.((((((((.......((((...)))).......))).)))))(((.((..(.(((....))))..)).)))..)))..)))))))))((((........)))).. ( -43.74) >DroVir_CAF1 6854 120 - 1 CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUUACGCGCAAGGAGCACCUCACGAAUCAUGUGCGACAGCACACGGGCGAU .((((((....)))))).((.(........).)).((((((......(((((.....))))).....(((.....((..(((....)))..))......((((....))))))))))))) ( -32.00) >DroGri_CAF1 7484 120 - 1 CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUAUUGCACCAAGACGUUUACGCGCAAGGAGCACCUCACCAAUCAUGUGCGACAGCAUACGGGCGAU .((((((....)))))).((.(........).)).((((((......(((((.....)))))......(((..(((((((((....))).........))))))..))).....)))))) ( -30.40) >DroWil_CAF1 10448 120 - 1 CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACCUAUUGCACCAAGACGUUCACACGCAAGGAGCAUCUGACGAAUCAUGUGCGCCAGCAUACUGGCGAC (((((((((..((.((((.(....).))))((((...))))......(((((.....)))))..(((.((((...(....))))).)))))..)))))))))((((((....)))))).. ( -42.00) >DroMoj_CAF1 7354 120 - 1 CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUUACGCGCAAGGAGCAUUUGACGAAUCAUGUGCGCCAGCACACGGGCGAU (((((((((..(((((((.(....).))))((((...))))(((....)))......(((.((..(.(((....))))..)).)))..)))..)))))))))((((........)))).. ( -37.30) >DroAna_CAF1 10784 120 - 1 CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAGUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUCACGCGCAAGGAGCAUUUGACGAAUCAUGUGCGCCAGCAUACGGGCGAC (((((((((..(((((((.(....).))))((((...))))(((....)))......(((.((..(.(((....))))..)).)))..)))..)))))))))((((........)))).. ( -37.30) >consensus CACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUCACGCGCAAGGAGCACCUGACGAAUCAUGUGCGCCAGCACACGGGCGAC (((((((((..(((((((.(....).))))((((...))))(((....)))......(((.((..(.(((....))))..)).)))..)))..)))))))))((((........)))).. (-33.38 = -33.77 + 0.39)

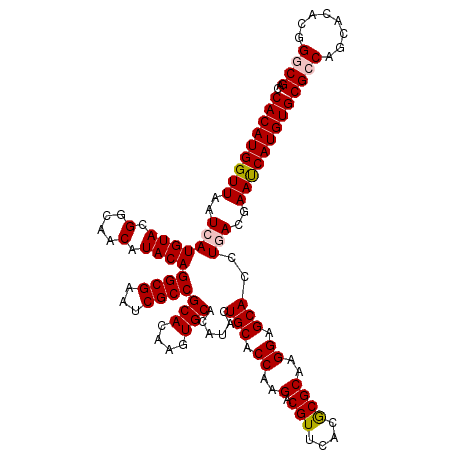
| Location | 11,799,101 – 11,799,221 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -44.57 |
| Consensus MFE | -44.48 |
| Energy contribution | -43.48 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.46 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.13 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11799101 120 + 22407834 UUGCGCGUGAACGUCUUGGUGCAGUACGUGCACUUGUGCGGCGACUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGCGUGAACGCUUUCGUGCAGUACGUGCACUUG .((((((((.(((....((((...((((((((..(((((((((((..........))))))))))).(((((......)))))..))))))))..))))..)))....)))))))).... ( -49.20) >DroVir_CAF1 6894 120 + 1 UUGCGCGUAAACGUCUUGGUGCAGUAUGUGCACUUGUGCGGCGAUUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGUGUGAACGCUUUCGUGCAGUACGUGCACUUG (..((((((((.(((.((((.(((((((.(((..(((((((((...)))).))))).)))))))).))....))))...))).))))))))..)........(((((.....)))))... ( -43.10) >DroGri_CAF1 7524 120 + 1 UUGCGCGUAAACGUCUUGGUGCAAUAUGUGCACUUGUGCGGCGAUUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGUGUGAACGCUUUCGUGCAGAACGUGCACUUG (..((((((((.(((.(((((((.....))).(.(((((((((((..........))))))))))).)....))))...))).))))))))..)........(((((.....)))))... ( -43.00) >DroWil_CAF1 10488 120 + 1 UUGCGUGUGAACGUCUUGGUGCAAUAGGUGCACUUGUGCGGCGAUUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGCGUGAACGCUUUUGUGCAGUACGUGCACUUG (..((((..((.(((.(((((((.....))).(.(((((((((((..........))))))))))).)....))))...))).))..))))..)........(((((.....)))))... ( -42.20) >DroMoj_CAF1 7394 120 + 1 UUGCGCGUAAACGUCUUGGUGCAGUAUGUGCACUUGUGCGGCGAUUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGUGUGAACGCUUUCGUGCAGUACGUGCACUUG (..((((((((.(((.((((.(((((((.(((..(((((((((...)))).))))).)))))))).))....))))...))).))))))))..)........(((((.....)))))... ( -43.10) >DroAna_CAF1 10824 120 + 1 UUGCGCGUGAACGUCUUGGUGCAGUAUGUGCACUUGUGCGGCGACUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGCGUAAACGCUUUCGUGCAGUAGGUGCACUUG (((((((..((.(((.(((((((.....))).(.(((((((((((..........))))))))))).)....))))...))).))..)))))))........(((((.....)))))... ( -46.80) >consensus UUGCGCGUAAACGUCUUGGUGCAGUAUGUGCACUUGUGCGGCGAUUCGCCUGUAUGUUGCCGUACAUGAUUAACCAUGUGAUCUUUGCGCGUGAACGCUUUCGUGCAGUACGUGCACUUG (((((((((((.(((.(((((((.....))).(.(((((((((((..........))))))))))).)....))))...))).)))))))))))........(((((.....)))))... (-44.48 = -43.48 + -1.00)

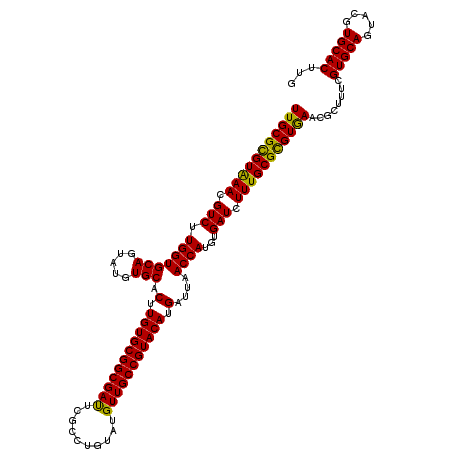
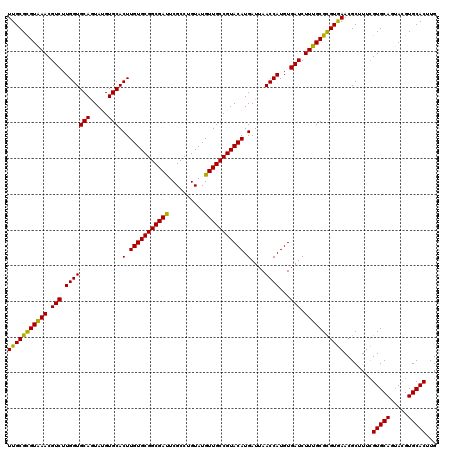
| Location | 11,799,101 – 11,799,221 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -31.57 |
| Energy contribution | -31.35 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11799101 120 - 22407834 CAAGUGCACGUACUGCACGAAAGCGUUCACGCGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAGUCGCCGCACAAGUGCACGUACUGCACCAAGACGUUCACGCGCAA ...(((((.(((((((((....(((....)))((.......((((((....))))))..(....).....((((...))))))....))))).)))))))))...(.(((....)))).. ( -38.30) >DroVir_CAF1 6894 120 - 1 CAAGUGCACGUACUGCACGAAAGCGUUCACACGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUUACGCGCAA ...((((..(((.(((.((...((((....)))).......((((((....)))))).)))))...))).((((...))))))))..(((((.....)))))...(.(((....)))).. ( -33.00) >DroGri_CAF1 7524 120 - 1 CAAGUGCACGUUCUGCACGAAAGCGUUCACACGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUAUUGCACCAAGACGUUUACGCGCAA ...((((((((..((..((...((((....)))).......((((((....)))))).))((((......((((...))))(((....))).....))))..))..))))....)))).. ( -32.60) >DroWil_CAF1 10488 120 - 1 CAAGUGCACGUACUGCACAAAAGCGUUCACGCGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACCUAUUGCACCAAGACGUUCACACGCAA ...(((((.....)))))....((((..((((((.......((((((....))))))..(....).....((((...))))))....(((((.....)))))...).)))....)))).. ( -31.90) >DroMoj_CAF1 7394 120 - 1 CAAGUGCACGUACUGCACGAAAGCGUUCACACGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUUACGCGCAA ...((((..(((.(((.((...((((....)))).......((((((....)))))).)))))...))).((((...))))))))..(((((.....)))))...(.(((....)))).. ( -33.00) >DroAna_CAF1 10824 120 - 1 CAAGUGCACCUACUGCACGAAAGCGUUUACGCGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAGUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUCACGCGCAA ...(((((.....)))))....((((..((((((.......((((((....))))))..(....).....((((...))))))....(((((.....)))))...).)))..)))).... ( -34.40) >consensus CAAGUGCACGUACUGCACGAAAGCGUUCACACGCAAAGAUCACAUGGUUAAUCAUGUACGGCAACAUACAGGCGAAUCGCCGCACAAGUGCACAUACUGCACCAAGACGUUCACGCGCAA ...((((......(((.((...((((....)))).......((((((....)))))).))))).......((((...))))))))..(((((.....)))))...(.(((....)))).. (-31.57 = -31.35 + -0.22)
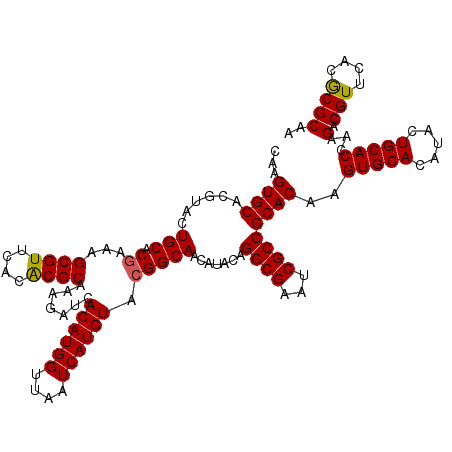
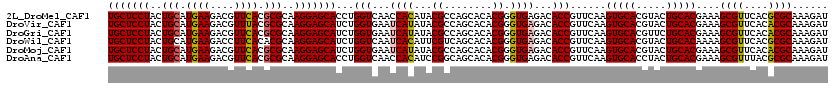
| Location | 11,799,181 – 11,799,301 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -33.56 |
| Energy contribution | -33.42 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11799181 120 - 22407834 UGCUCCUACUGCAUGAAGACGUUCACGCGCAAGGAGCACCUGGUCAACCACAUACGCCAGCACACGGGUGAGACACCGUUCAAGUGCACGUACUGCACGAAAGCGUUCACGCGCAAAGAU (((((((..(((.((((....)))).)))..))))))).(((((...........)))))....((.(((((....((((...(((((.....)))))...))))))))).))....... ( -36.70) >DroVir_CAF1 6974 120 - 1 UGCUCCUACUGCAUGAAGACGUUUACGCGCAAGGAGCAUCUGGUGAAUCAUAUACGCCAGCACACGGGUGAGACACCGUUCAAGUGCACGUACUGCACGAAAGCGUUCACACGCAAAGAU (((((((..(((.((((....)))).)))..)))))))(((((((..((((...((........)).))))..))))......(((((.....)))))....((((....))))..))). ( -36.00) >DroGri_CAF1 7604 120 - 1 UGCUCCUACUGCAUGAAGACGUUCACGCGCAAGGAGCAUCUGGUGAAUCAUAUACGCCAGCACACGGGUGAGACACCGUUCAAGUGCACGUUCUGCACGAAAGCGUUCACACGCAAAGAU (((((((..(((.((((....)))).)))..)))))))(((((((..((((...((........)).))))..))))......(((((.....)))))....((((....))))..))). ( -38.20) >DroWil_CAF1 10568 120 - 1 UGCUCCUACUGCAUGAAGACCUUCACACGCAAGGAGCAUCUGGUCAAUCACAUUCGUCAGCACACGGGUGAGACACCGUUCAAGUGCACGUACUGCACAAAAGCGUUCACGCGCAAAGAU (((((((..(((.((((....))))...))))))))))((((((...((.(((((((......))))))).)).)))......(((((.....)))))....((((....))))..))). ( -36.10) >DroMoj_CAF1 7474 120 - 1 UGCUCCUACUGCAUGAAGACGUUCACGCGCAAGGAGCAUCUGGUGAAUCAUAUACGCCAGCACACGGGUGAGACACCGUUCAAGUGCACGUACUGCACGAAAGCGUUCACACGCAAAGAU (((((((..(((.((((....)))).)))..)))))))(((((((..((((...((........)).))))..))))......(((((.....)))))....((((....))))..))). ( -38.20) >DroAna_CAF1 10904 120 - 1 UGCUCCUACUGCAUGAAGACGUUCACGCGCAAGGAGCACCUGGUCAACCACAUCCGGCAGCACACGGGUGAGACACCGUUCAAGUGCACCUACUGCACGAAAGCGUUUACGCGCAAAGAU (((((((..(((.((((....)))).)))..)))))))..(((....)))((((((........)))))).............(((((.....)))))....((((....))))...... ( -37.90) >consensus UGCUCCUACUGCAUGAAGACGUUCACGCGCAAGGAGCAUCUGGUCAAUCACAUACGCCAGCACACGGGUGAGACACCGUUCAAGUGCACGUACUGCACGAAAGCGUUCACACGCAAAGAU (((((((..(((.((((....)))).)))..)))))))...(((...((((...((........)).))))...)))......(((((.....)))))....((((....))))...... (-33.56 = -33.42 + -0.14)
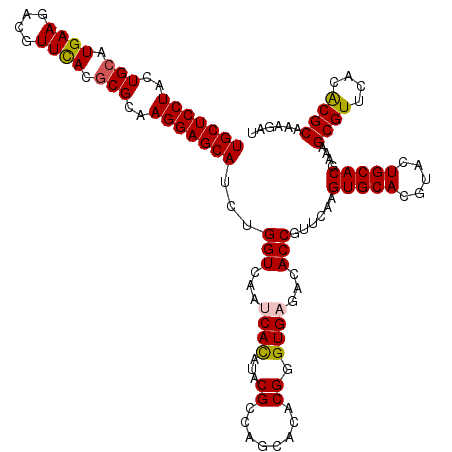
| Location | 11,799,221 – 11,799,341 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -49.28 |
| Consensus MFE | -41.21 |
| Energy contribution | -41.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11799221 120 + 22407834 AACGGUGUCUCACCCGUGUGCUGGCGUAUGUGGUUGACCAGGUGCUCCUUGCGCGUGAACGUCUUCAUGCAGUAGGAGCAGCGGUGUGGCGACUCUCCCGUAUGCUGGCGCACGUGGUUA ...........((((((((((..(((((((.(((((.((((.(((((((.(((((((((....))))))).))))))))).)....))))))))....)))))))..))))))).))).. ( -58.20) >DroVir_CAF1 7014 120 + 1 AACGGUGUCUCACCCGUGUGCUGGCGUAUAUGAUUCACCAGAUGCUCCUUGCGCGUAAACGUCUUCAUGCAGUAGGAGCAGCGAUGCGGCGACUCUCCCGUGUGCUGGCGCACGUGAUUU ...((((...))))(((((((..((((((..((.((.((...(((((((.((((((..........)))).)))))))))((...)))).)).))....))))))..)))))))...... ( -43.10) >DroGri_CAF1 7644 120 + 1 AACGGUGUCUCACCCGUGUGCUGGCGUAUAUGAUUCACCAGAUGCUCCUUGCGCGUGAACGUCUUCAUGCAGUAGGAGCAGCGAUGCGGCGACUCUCCCGUGUGCUGGCGCACAUGAUUU ...((((...)))).((((((..((((((..((.((.((...(((((((.(((((((((....))))))).)))))))))((...)))).)).))....))))))..))))))....... ( -48.00) >DroWil_CAF1 10608 120 + 1 AACGGUGUCUCACCCGUGUGCUGACGAAUGUGAUUGACCAGAUGCUCCUUGCGUGUGAAGGUCUUCAUGCAGUAGGAGCAGCGAUGGGGCGAUUCACCCGUGUGCUGACGCACGUGAUUU ...((((..((.(((((.((((.....((.((......)).))((((((.(((((((((....))))))).)))))))))))))))))..))..))))((((((....))))))...... ( -44.50) >DroMoj_CAF1 7514 120 + 1 AACGGUGUCUCACCCGUGUGCUGGCGUAUAUGAUUCACCAGAUGCUCCUUGCGCGUGAACGUCUUCAUGCAGUAGGAGCAGCGAUGCGGCGACUCUCCCGUGUGCUGCCGCACAUGAUUU ...((((...))))(((((((.(((..................((((((.(((((((((....))))))).))))))))(((.((((((.(.....)))))))))))))))))))).... ( -48.50) >DroAna_CAF1 10944 120 + 1 AACGGUGUCUCACCCGUGUGCUGCCGGAUGUGGUUGACCAGGUGCUCCUUGCGCGUGAACGUCUUCAUGCAGUAGGAGCAGCGGUGCGGCGACUCUCCCGUGUGCUGGCGCACGUGGUUG ...........((((((((((((((((..((.(((((((.(.(((((((.(((((((((....))))))).))))))))).)))).)))).))....)))...)).)))))))).))).. ( -53.40) >consensus AACGGUGUCUCACCCGUGUGCUGGCGUAUAUGAUUCACCAGAUGCUCCUUGCGCGUGAACGUCUUCAUGCAGUAGGAGCAGCGAUGCGGCGACUCUCCCGUGUGCUGGCGCACGUGAUUU ...((((...)))).((((((((((...............(.(((((((.(((((((((....))))))).))))))))).).((((((.(.....)))))))))))))))))....... (-41.21 = -41.60 + 0.39)

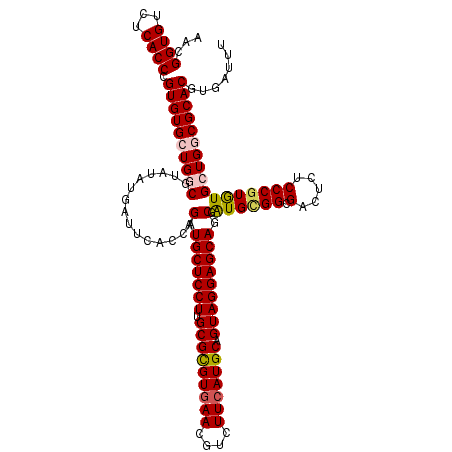
| Location | 11,799,341 – 11,799,455 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.71 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -25.56 |
| Energy contribution | -25.53 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11799341 114 + 22407834 AGCAAGUGCUCCUUUCGCGUAAACGUCUUGGAGCAAAAGUCGCACCGGUGCGGCGUCUCGCCUGCUUGCGUGGCAAAUGUGUGGGUGUCGCGCGAAUGGGACGAGAA----AAU-GUUU ((((..((((((....(((....)))...))))))...((((((....)))))).((((((((..((((((((((..(....)..))))))))))..))).))))).----..)-))). ( -49.40) >DroVir_CAF1 7134 101 + 1 AACAAAUGCUCCUUGCGCGUAAACGUCUUGGAGCAGAAGUCGCACCGAUGCGGCGUCUCGCCUGCUUGCGUGGUAAAAG-GUAA---------GAAUCGUACAAUAA----CA----UU .((...((((((..(.(((....))))..))))))((.((((((....)))))).(((..(((..((((...)))).))-)..)---------)).)))).......----..----.. ( -30.40) >DroPse_CAF1 10756 105 + 1 AACAAAUGCUCCUUGCGCGUAAACGUCUUGGAGCAGAAGUCGCACCGAUGCGGCGUCUCGCCUGCUUGCGUGGCAAAAGUGUGAG---------GAGAUAUCAAGAA----AAU-GUUU ........(((((..(((.....(((.((((.((.......)).)))).)))..(((.(((......))).)))....)))..))---------)))..........----...-.... ( -33.40) >DroWil_CAF1 10728 104 + 1 AACAGAUGUUCCUUGCGCGUAAACGUCUUGGAGCAGAAAUCGCAACGAUGCGGCGUCUCGCCUGCUUGCAUGGCAA-UGAG------------AAAU--GACAAAAUAAUAAAUAUUUU ......((((((..(.(((....))))..))))))....(((((....)))))..(((((..((((.....)))).-))))------------)...--.................... ( -23.60) >DroAna_CAF1 11064 102 + 1 AGCAAGUGUUCCUUGCGCGUAAACGUCUUGGAGCAGAAGUCGCACCGGUGCGGCGUCUCGCCUGCUUGCGUGGCAAAUGAUUCG------A-------GAGCAAGAA----AAUUUUUU .((...((((((..(.(((....))))..))))))(((((((((....))))))(((.(((......))).)))......))).------.-------..)).....----........ ( -31.70) >DroPer_CAF1 10858 105 + 1 AACAAAUGCUCCUUGCGCGUAAACGUCUUGGAGCAGAAGUCGCACCGAUGCGGCGUCUCGCCUGCUUGCGUGGCAAAAGUGUGAG---------GAGAUAUCAAGAA----AAU-GUUU ........(((((..(((.....(((.((((.((.......)).)))).)))..(((.(((......))).)))....)))..))---------)))..........----...-.... ( -33.40) >consensus AACAAAUGCUCCUUGCGCGUAAACGUCUUGGAGCAGAAGUCGCACCGAUGCGGCGUCUCGCCUGCUUGCGUGGCAAAAGUGUGA__________AAU_GAACAAGAA____AAU_GUUU ......((((((....(((....)))...))))))...((((((....))))))(((.(((......))).)))............................................. (-25.56 = -25.53 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:41 2006