
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,793,627 – 11,793,747 |
| Length | 120 |
| Max. P | 0.883484 |

| Location | 11,793,627 – 11,793,747 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -47.43 |
| Consensus MFE | -28.67 |
| Energy contribution | -28.37 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11793627 120 + 22407834 AAAGAGUGAAUCCUCUCCAGCGUGUGGUUACUGAGCAGUCGCAGUCUUCGGACAUCCAACUGCUUGCAACUCAGGUGCUGCAUGUGCGGAGCCAGGGCCAAGAUGAAAGCCACAAACUGU ..((((......)))).(((..(((((((...(((((((.(..(((....)))...).)))))))....(((.(((.((((....)))).))).)))..........)))))))..))). ( -40.50) >DroPse_CAF1 4089 120 + 1 AGGCUGGUCACCCCGGACAGGGUAUGGCUGCUCAACAGGCGAAGCCGCCUCACAUCCAGCCGCUGGCAGCUCAAACGCUGCAUGUGCGGUGCCAGGGUAAGCACAAAGGCCAGAAACUGA ((.((((((((((......))))..(((((......(((((....)))))......)))))....(((((......))))).(((((..(((....))).)))))..))))))...)).. ( -49.70) >DroSim_CAF1 992 120 + 1 AAGGAGUGAAUCCUCUCCAGGGUGUGGUUACUGAGCAGUCGCAGUCUUCGGAGAUCCAACUGCUUGCAGCUCAGGCGCUGCAUAUGCGGAGCCAGGGCCAGAAUGAAAGCCACAAACUGU ..((((.(....).))))((..(((((((...(((((((.(.(.((....))..).).)))))))...((((.(((.((((....)))).))).)))).........)))))))..)).. ( -45.80) >DroEre_CAF1 966 120 + 1 AAGGAGUGAAUCCUCUCCAGGGUGUGGCUACUAAGCACCCGCAGCCGGCGGACAUCCAACUGCUGGCAGCUCAGGCGCUGCAUGUGCGGAGCCAGGGCCAAAAUGAAAGCCUCAAAUUGU ..((((.(....).)))).((((((.........))))))((.((((((((........)))))))).((((.(((.((((....)))).))).))))..........)).......... ( -48.70) >DroYak_CAF1 980 120 + 1 AAGGAGUGAAUGCUCUCCAGGGUGUGGCUACUAAGCAGCCGCAGUCUUCGGACAUCCAACUGCUGGCAGCUCAGGCGCUGCAUGUGCGGAGCCAGGGCCUGGAUGAAAGCAACAAACUGU ....(((...((((.((((((.(.(((((.((.(((.((((((((....((....)).))))).))).))).))((((.....))))..))))).).))))))....))))....))).. ( -49.20) >DroPer_CAF1 4085 120 + 1 AGGCUGGUCACCCCGGACAGGGUAUGGCUGCUCAACAGGCGAAGCCGCCUCACAUCCAGCCGCUGGCAGCUCAAACGCUGCAUGUGCGGUGCCAGGGUCAGCACAAAGGCCAGAAACUGA ((.((((((((((......)))).(((((((((...(((((....)))))........(((((..(((((......)))))....)))))....))).)))).))..))))))...)).. ( -50.70) >consensus AAGGAGUGAAUCCUCUCCAGGGUGUGGCUACUAAGCAGCCGCAGCCGUCGGACAUCCAACUGCUGGCAGCUCAGGCGCUGCAUGUGCGGAGCCAGGGCCAGAAUGAAAGCCACAAACUGU ...(((......)))..(((..(((((((..............................((((..(((((......)))))....)))).((....)).........)))))))..))). (-28.67 = -28.37 + -0.30)

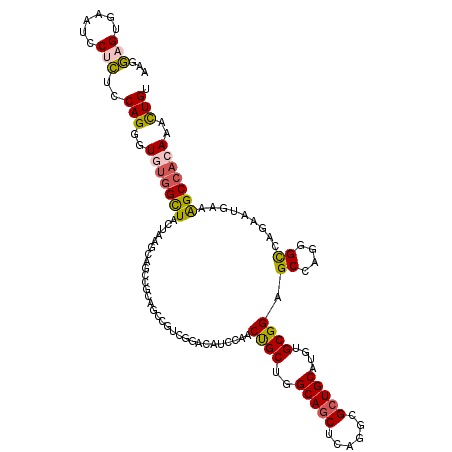
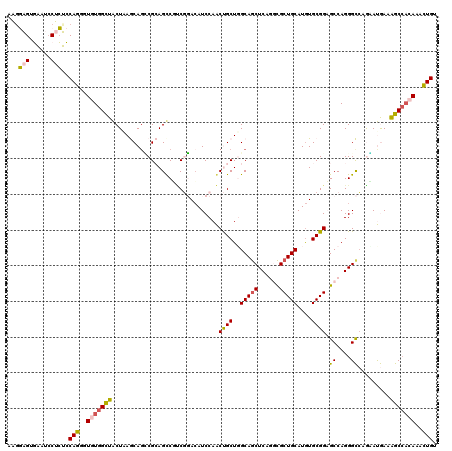
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:28 2006