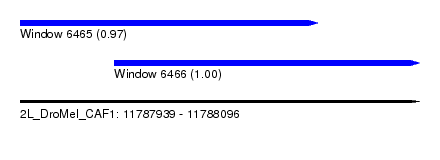
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,787,939 – 11,788,096 |
| Length | 157 |
| Max. P | 0.999982 |

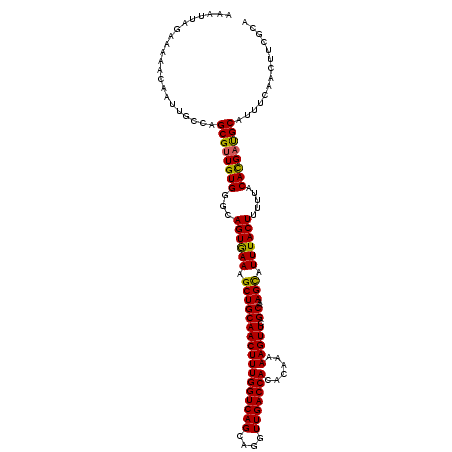
| Location | 11,787,939 – 11,788,056 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.51 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -24.10 |
| Energy contribution | -25.34 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11787939 117 + 22407834 GCAAACGAAAUGCAAAGAGACGCAUAUUUUU---UUACUUAAAUUAGAAAAACAAUUGCCAGCGUUGUGGGUAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCG ((((.....((((........)))).(((((---(((.......))))))))...)))).(((.(((((.(((((....))))).....(((((((....))))))))))))...))).. ( -29.70) >DroSec_CAF1 45656 97 + 1 -------AAAUGCAAAGA-ACGCAUAUUUUU---UUACUUAAA------------UUGCCAGCGCUGUGGGCAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCG -------.........((-(((((.((((..---......)))------------))))......((((.(((((....))))).....(((((((....)))))))))))....)))). ( -28.30) >DroSim_CAF1 44860 117 + 1 GCAAACGAAAUGCAAAGAGACGCAUAUUUUU---UUACUUAAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCG ((((.....((((........)))).(((((---(((.......))))))))...)))).(((.(((((.(((((....))))).....(((((((....))))))))))))...))).. ( -31.70) >DroEre_CAF1 56076 120 + 1 GCAAACGAAAUGCAAAGAGACGCAUAUUUUUUUUUUACUUAAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUAAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCG ((((.....((((........))))....((((((((.......))))))))...)))).(((.(((((.(((((....))))).....(((((((....))))))))))))...))).. ( -29.30) >DroYak_CAF1 47762 120 + 1 GCAAACGAAAUGCAAAGAGACGCAUAUUUUUUUGUUACUUAAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUAAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCG ((((.....((((........)))).((((((((..........))))))))...)))).(((.(((((.(((((....))))).....(((((((....))))))))))))...))).. ( -29.40) >consensus GCAAACGAAAUGCAAAGAGACGCAUAUUUUU___UUACUUAAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCG .........((((........))))...................................(((.(((((.(((((....))))).....(((((((....))))))))))))...))).. (-24.10 = -25.34 + 1.24)

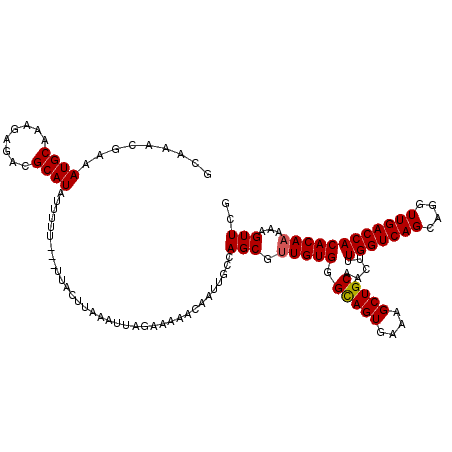
| Location | 11,787,976 – 11,788,096 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -31.66 |
| Energy contribution | -31.06 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.28 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11787976 120 + 22407834 AAAUUAGAAAAACAAUUGCCAGCGUUGUGGGUAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCGCAAAGUAUUUACUUUUUUCACGAUGCAUUUCAACUUCGCA .....................((((((((((.((((((.(((((((((((((((((....)))))))......))))).))..))).))))))...)))))))))).............. ( -32.50) >DroSec_CAF1 45685 108 + 1 AAA------------UUGCCAGCGCUGUGGGCAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCGCAAAGCAUUUACUUUUUACACGAUGCAUUUUAACUUCGCA ...------------((((.(((..((((.(((((....))))).....(((((((....)))))))))))....))).)))).(((((............))))).............. ( -31.00) >DroSim_CAF1 44897 120 + 1 AAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCGCAAAGCAUUUACUUUUUACACGAUGCAUUUCAACUUCGCA ......((((.....((((.(((.(((((.(((((....))))).....(((((((....))))))))))))...))).)))).(((((............))))).))))......... ( -35.40) >DroEre_CAF1 56116 120 + 1 AAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUAAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCGCAAAGCAUUUACUUUUUACAUGACGCAUUUCAACUUCGCA .....................((((..((...((((((.(((((((((((((((((....)))))))......))))).))..))).)))))).....))..)))).............. ( -31.10) >DroYak_CAF1 47802 120 + 1 AAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUAAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCGCAAAGCAUUUACUUUUUACACGACGCAUUUCAACUUCGCA .....................((((((((...((((((.(((((((((((((((((....)))))))......))))).))..))).)))))).....)))))))).............. ( -33.50) >consensus AAAUUAGAAAAACAAUUGCCAGCGUUGUGGGCAGUGAAAGCUGCAACUUUGGUCAGCAGGUUGACCACACAAAAAGUUCGCAAAGCAUUUACUUUUUACACGAUGCAUUUCAACUUCGCA .....................((((((((...((((((.(((((((((((((((((....)))))))......))))).))..))).)))))).....)))))))).............. (-31.66 = -31.06 + -0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:26 2006