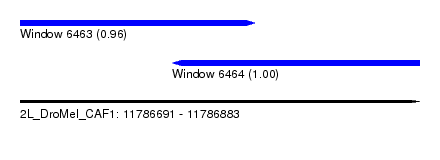
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,786,691 – 11,786,883 |
| Length | 192 |
| Max. P | 0.995573 |

| Location | 11,786,691 – 11,786,804 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.46 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -28.28 |
| Energy contribution | -28.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
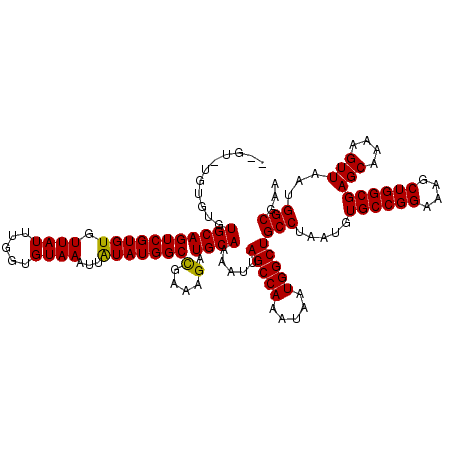
>2L_DroMel_CAF1 11786691 113 + 22407834 -------UGUGUGCAGUCGUGUGUUAUUUGGUGUAAAUUAUAUGGCCGAAAGAUGCAAAAUUAGCCAAAUAAUGGCUGCCUAAUGUGCCGGAAAGCUGGCGAGCAAAAGUUAAUGGCGAA -------....((((((((((((((((.....))))..))))))))(....).)))).....(((((.....)))))(((.....((((((....))))))(((....)))...)))... ( -29.70) >DroSec_CAF1 44441 118 + 1 --GUGUGUGUGUGCAGUCGUGUGUUAUUUGGUGUAAAUUAUAUGGCCGAAAGAUGCAAAAUUAGCCAAAUAAUGGCUGCCUAAUGUGCCGGAAAGCUGGCGAGCAAAAGUUAAUGGCGAA --.........((((((((((((((((.....))))..))))))))(....).)))).....(((((.....)))))(((.....((((((....))))))(((....)))...)))... ( -29.70) >DroSim_CAF1 43648 116 + 1 --G--UGUGUGUGCAGUCGUGUGUUAUUUGGUGUAAAUUAUAUGGCCGAAAGAUGCAAAAUUAGCCAAAUAAUGGCUGCCUAAUGUGCCGGAAAGCUGGCGAGCAAAAGUUAAUGGCGAA --.--......((((((((((((((((.....))))..))))))))(....).)))).....(((((.....)))))(((.....((((((....))))))(((....)))...)))... ( -29.70) >DroEre_CAF1 54850 120 + 1 GGGUAUGUGUGUGCAGUCGUGUGUUAUUUGGUGUAAAUUGUAUGGCCGAAAGAUGCAAAAUUAGCCAAAUAAUGGCUGCCUAAUGUGCCGGAAAGCUGGCGAGCAAAAGUUAAUGGCGAA .(((((((....((((((((....((((((((.....((((((...(....))))))).....))))))))))))))))...))))))).....((((...(((....)))..))))... ( -35.90) >DroYak_CAF1 46494 118 + 1 --GUGUGUGUGUGCAGUCGUGUGUUAUUUGGUGUAAAUUAUAUGGCUGAAAGAUGCAAAAUUAGCCAAAUAAUGGCUGCCUAAUGUGCCGGAAAGCUGGCGAGCAAAAGUUAAUGGCGAA --.(((((.(...((((((((((((((.....))))..))))))))))..).))))).....(((((.....)))))(((.....((((((....))))))(((....)))...)))... ( -33.60) >consensus __GU_UGUGUGUGCAGUCGUGUGUUAUUUGGUGUAAAUUAUAUGGCCGAAAGAUGCAAAAUUAGCCAAAUAAUGGCUGCCUAAUGUGCCGGAAAGCUGGCGAGCAAAAGUUAAUGGCGAA ...........(((((((((((.((((.....))))...)))))))(....).)))).....(((((.....)))))(((.....((((((....))))))(((....)))...)))... (-28.28 = -28.36 + 0.08)


| Location | 11,786,764 – 11,786,883 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.41 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -32.10 |
| Energy contribution | -31.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
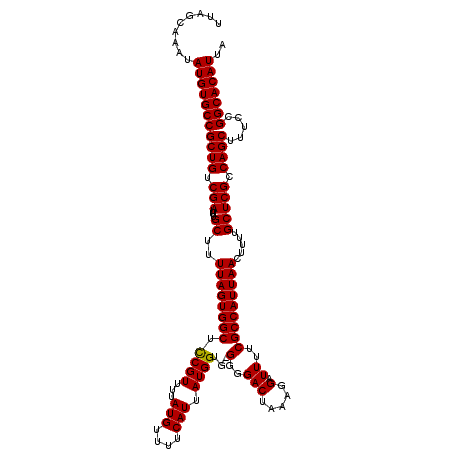
>2L_DroMel_CAF1 11786764 119 - 22407834 UUAGCAAAUAUGUGCCGCUGUCGAUUUGCUUUUAGUGGCUCCGUUUUAUGUUUUCAUUAUGGUG-GGGGACUAAAGGAUUUUCGCCAUUAACUUUUGCUCGCCAGCUUUCCGGCACAUUA .........(((((((((((.(((...((..((((((((.((((...(((....))).))))((-(....)))..........)))))))).....))))).)))).....))))))).. ( -34.50) >DroSec_CAF1 44519 119 - 1 UUAGCAAAUAUGUGCCGCUGUCGAUUUGCUUUUAGUGGCUCCGUUUUAUGUUUUCAUUAUGGUG-GGGGACUAAAGGAUUUUCGCCAUUAACUUUUGCUCGCCAGCUUUCCGGCACAUUA .........(((((((((((.(((...((..((((((((.((((...(((....))).))))((-(....)))..........)))))))).....))))).)))).....))))))).. ( -34.50) >DroSim_CAF1 43724 119 - 1 UUAGCAAAUAUGUGCCGCUGUCGAUUUGCUUUUAGUGGCUCCGUUUUAUGUUUUCAUUAUGGUG-GGGGACUAAAGGAUUUUCGCCAUUAACUUUUGCUCGCCAGCUUUCCGGCACAUUA .........(((((((((((.(((...((..((((((((.((((...(((....))).))))((-(....)))..........)))))))).....))))).)))).....))))))).. ( -34.50) >DroEre_CAF1 54930 119 - 1 UUAGCAAAUAUGUGCCGCUGUCGAUUUGCUUUUAGUGGCUCCGUUUUAUGUUUUCAUUAUGGAG-GGGGACUAAAGGAUUUUCGCCAUUAACUUUUGCUCGCCAGCUUUCCGGCACAUUA .........(((((((((((.(((...((..(((((((((((((...(((....))).)))))(-(....))...........)))))))).....))))).)))).....))))))).. ( -35.60) >DroYak_CAF1 46572 120 - 1 UUAGCAACUAUGUGCCGCUGUCGAUUUGCUUUUAGUGGCUUCGUUUUAUGUUUUCAUUAUGGGGAGGGGACUAAAGGAUUUUCGCCAUUAACUUUUGCUCGCCAGCUUUCCGGCACAUUA .........(((((((((((.(((...((..(((((((((((((...(((....))).)))))(((((..(....)..))))))))))))).....))))).)))).....))))))).. ( -32.40) >consensus UUAGCAAAUAUGUGCCGCUGUCGAUUUGCUUUUAGUGGCUCCGUUUUAUGUUUUCAUUAUGGUG_GGGGACUAAAGGAUUUUCGCCAUUAACUUUUGCUCGCCAGCUUUCCGGCACAUUA .........(((((((((((.(((...((..((((((((.((((...(((....))).))))...(..(((.....).))..))))))))).....))))).)))).....))))))).. (-32.10 = -31.94 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:24 2006