
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
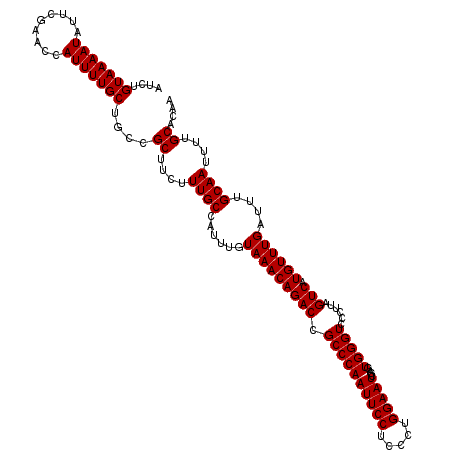
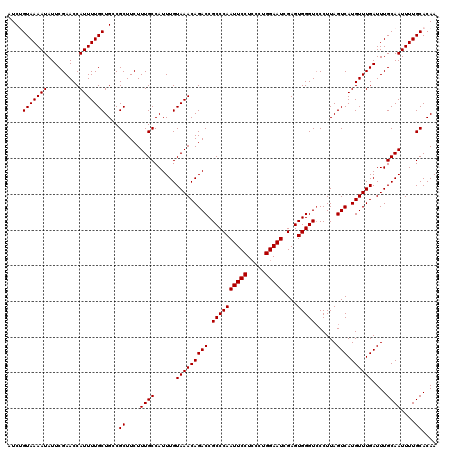
| Location | 11,785,932 – 11,786,132 |
| Length | 200 |
| Max. P | 0.998562 |

| Location | 11,785,932 – 11,786,052 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11785932 120 + 22407834 AUCUGUAAAAUAUUCGAACCAUUUUGCUGCCGCUUCUUUGCCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAA ....(((((((.........)))))))....((....((((......(((((((((.((((((((((.....)))))....)))))......))).))))))....))))....)).... ( -24.80) >DroSec_CAF1 43678 120 + 1 AUCUGUAAAAUAUUCGAACCAUUUUGCUGGCGCUUCUUUGCCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUGCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAA ...((((((((..((((((...((((((((((......)))))...)))))..((((((((((((((.....)))))....)))))).....)))..))))))......))))))))... ( -30.30) >DroSim_CAF1 42887 120 + 1 AUCUGUAAAAUAUUCGAACCAUUUUGCUGGCGCUUCUUUGCCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAA ...((((((((..((((((...((((((((((......)))))...)))))..(((.((((((((((.....)))))....)))))......)))..))))))......))))))))... ( -27.50) >DroEre_CAF1 54089 120 + 1 AUCAGUAAAAUAUUCGAACCAUUUUGCUGCCGCUUCUUUGCCAUUUGUAAACAGACCGCCCAAUUCCUAUCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAA ..(((((((((.........)))))))))..((....((((......(((((((((.((((((((((.....)))))....)))))......))).))))))....))))....)).... ( -28.30) >DroYak_CAF1 45715 120 + 1 AUCAGUAAAAUAUUCGAACCAUUUUGCUGCCGCUUCUUUGCCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAA ..(((((((((.........)))))))))..((....((((......(((((((((.((((((((((.....)))))....)))))......))).))))))....))))....)).... ( -28.30) >consensus AUCUGUAAAAUAUUCGAACCAUUUUGCUGCCGCUUCUUUGCCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAA ....(((((((.........)))))))....((....((((......(((((((((.((((((((((.....)))))....)))))......))).))))))....))))....)).... (-24.88 = -24.88 + 0.00)

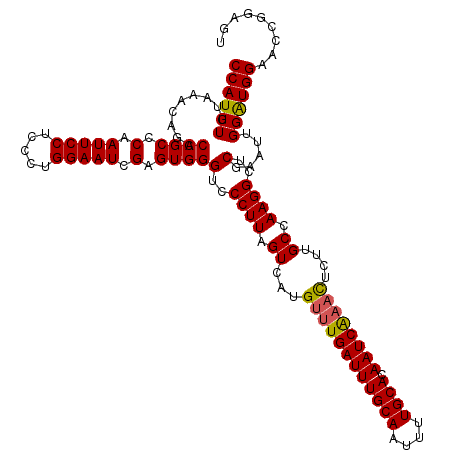
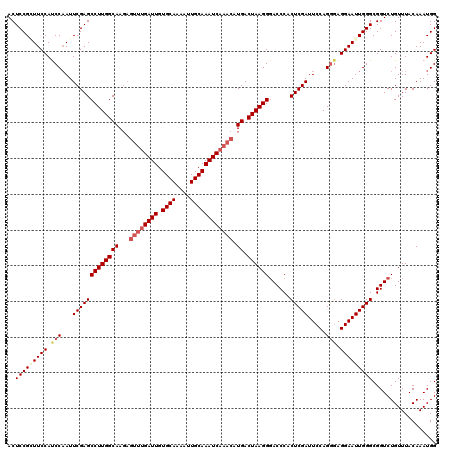
| Location | 11,785,972 – 11,786,092 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -31.72 |
| Energy contribution | -31.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11785972 120 + 22407834 CCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAAUCGUUUGAUUGCCAAGGCUCGAAUGGGAUGGAAUCGGAGU ((.((((....))))(((((((.((((.....))))((((((.(((.....(((((....(((((((((....)))).)))))..))))))))...)))))).)))).)))....))... ( -35.10) >DroSec_CAF1 43718 120 + 1 CCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUGCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAAUCAAACUCUUGCCAAGGCUCGAAUUGGAUGGAAGCGGAGU ((((((.........((((.(.(((((.....))))).).))))(.(((((.((...((((((((((((....)))).))))))))....)).))))).).....))))))......... ( -37.20) >DroSim_CAF1 42927 120 + 1 CCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAAUCAAACUCUUGCCAAGGCUCGAAUUGGAUGGAAGCGGAGU ...((((....))))(((((((.((((.....))))((((((.(((.....((....((((((((((((....)))).)))))))).)).)))...))))))......)))..))))... ( -34.60) >DroEre_CAF1 54129 120 + 1 CCAUUUGUAAACAGACCGCCCAAUUCCUAUCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAAUCAAACUCUUGCCAAGGCUCGAAUUGGGUGGAACCGGAGU ((.((((....))))((((((((((((.....))))((((((.(((.....((....((((((((((((....)))).)))))))).)).)))...)))))).))))))))....))... ( -41.30) >DroYak_CAF1 45755 120 + 1 CCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAAUCAAACUCUUGCCAAGGCUCGAAUUGGGUGGAACCGGAGU ((.((((....))))((((((((((((.....))))((((((.(((.....((....((((((((((((....)))).)))))))).)).)))...)))))).))))))))....))... ( -41.30) >consensus CCAUUUGUAAACAGACCGCCCAAUUCCUCCCUGGAAUCGAGUGGGUCCCUUAGUCAUGUUUGAUUUGCAAUUUUGCACAAUCAAACUCUUGCCAAGGCUCGAAUUGGAUGGAACCGGAGU ((((((.........((((.(.(((((.....))))).).))))(..((((.((...((((((((((((....)))).))))))))....)).))))..).....))))))......... (-31.72 = -31.56 + -0.16)


| Location | 11,785,972 – 11,786,092 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -38.16 |
| Consensus MFE | -33.78 |
| Energy contribution | -35.18 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11785972 120 - 22407834 ACUCCGAUUCCAUCCCAUUCGAGCCUUGGCAAUCAAACGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCACUCGAUUCCAGGGAGGAAUUGGGCGGUCUGUUUACAAAUGG ..(((((((((.((((..((((((((((((((((....)))))((((....))))............))))))......))))).....))))))))))))).................. ( -35.50) >DroSec_CAF1 43718 120 - 1 ACUCCGCUUCCAUCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGCACCCACUCGAUUCCAGGGAGGAAUUGGGCGGUCUGUUUACAAAUGG .........((((.........(((((((((...((((((((.((((....)))))))))))).)).)))))))..((.(((((((((.....))))))))).))...........)))) ( -39.30) >DroSim_CAF1 42927 120 - 1 ACUCCGCUUCCAUCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCACUCGAUUCCAGGGAGGAAUUGGGCGGUCUGUUUACAAAUGG .........((((..........((((((((...((((((((.((((....)))))))))))).)).))))))((((..(((((((((.....))))))))).)))).........)))) ( -37.80) >DroEre_CAF1 54129 120 - 1 ACUCCGGUUCCACCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCACUCGAUUCCAGAUAGGAAUUGGGCGGUCUGUUUACAAAUGG ....(((..((.(((((.(((((((((((((...((((((((.((((....)))))))))))).)).))))))......)))))((((.....))))))))).)).)))........... ( -38.00) >DroYak_CAF1 45755 120 - 1 ACUCCGGUUCCACCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCACUCGAUUCCAGGGAGGAAUUGGGCGGUCUGUUUACAAAUGG ..(((((((((.(((...(((((((((((((...((((((((.((((....)))))))))))).)).))))))......))))).....))).))))))))).................. ( -40.20) >consensus ACUCCGCUUCCAUCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCACUCGAUUCCAGGGAGGAAUUGGGCGGUCUGUUUACAAAUGG ..(((((((((.(((...(((((((((((((...((((((((.((((....)))))))))))).)).))))))......))))).....))).))))))))).................. (-33.78 = -35.18 + 1.40)


| Location | 11,786,012 – 11,786,132 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -26.50 |
| Energy contribution | -27.62 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.852046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11786012 120 - 22407834 CAAUUUUUGGGAUUUAGCCUGCUGGCUGGCUGCUCCCGAAACUCCGAUUCCAUCCCAUUCGAGCCUUGGCAAUCAAACGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCAC ....((((((((..(((((........))))).))))))))...........((((......((....))........((((.((((....))))))))............))))..... ( -31.20) >DroSec_CAF1 43758 116 - 1 CAAUUUUUGGGAUUUAGCCUGCUG----GCUGCUCCCGAAACUCCGCUUCCAUCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGCACCCAC ....((((((((..(((((....)----)))).)))))))).....................(((((((((...((((((((.((((....)))))))))))).)).)))))))...... ( -39.10) >DroSim_CAF1 42967 116 - 1 CAAUUUUUGGGAUUUAGCCUGCUG----GCUGCUCCCGAAACUCCGCUUCCAUCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCAC ....((((((((..(((((....)----)))).)))))))).(((((((...........))))(((((((...((((((((.((((....)))))))))))).)).))))))))..... ( -36.40) >DroEre_CAF1 54169 116 - 1 CAAUUUUUGGGAUUUAGCCUGCUG----GCUGCUCCUGAAACUCCGGUUCCACCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCAC ....(((..(((..(((((....)----)))).)))..)))....((((((.......((..((....))..))((((((((.((((....))))))))))))........))))))... ( -38.80) >DroYak_CAF1 45795 116 - 1 CAAUUUUUGGGAUUUAGUCUGCUG----GCAGCUCCUGAAACUCCGGUUCCACCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCAC .......((((.((((((((((..----(..((((..(((.....((.....))...))))))).)..)))...((((((((.((((....))))))))))))..)))))))...)))). ( -34.90) >consensus CAAUUUUUGGGAUUUAGCCUGCUG____GCUGCUCCCGAAACUCCGCUUCCAUCCAAUUCGAGCCUUGGCAAGAGUUUGAUUGUGCAAAAUUGCAAAUCAAACAUGACUAAGGGACCCAC ....((((((((..((((..........)))).))))))))(((.((((......)))).)))((((((((...((((((((.((((....)))))))))))).)).))))))....... (-26.50 = -27.62 + 1.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:22 2006