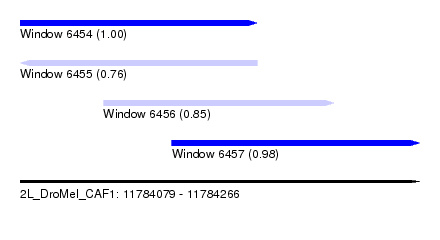
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,784,079 – 11,784,266 |
| Length | 187 |
| Max. P | 0.995310 |

| Location | 11,784,079 – 11,784,190 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -5.12 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11784079 111 + 22407834 AAAACAGCAACAAUAAUGUGUUAAUGUGUGCAAACACACACAAAUGCAUC----UGCAUACAUG----UAGAGUAUAGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUU ...............(((((((..((((((......))))))..((((((----(((((((...----....)))).((....)))))))...)))).))))))).............. ( -30.60) >DroSec_CAF1 41846 112 + 1 AAAACAGCAACAAUAAUGUGUUAAUGUGUGCAAACACACACAAAUGCAUC----UGCAUACAUACA--UAGAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAACGAGAAAUU ...............(((((((..((((((......))))))..((((((----(((.....(((.--....)))-.((....)))))))...)))).))))))).............. ( -29.90) >DroSim_CAF1 40950 112 + 1 AAAACAGCAACAAUAAUGUGUUAAUGUGUGCAAACACACACAAAUGCAUC----UGCAUACAUACA--UAGAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUU ...............(((((((..((((((......))))))..((((((----(((.....(((.--....)))-.((....)))))))...)))).))))))).............. ( -29.90) >DroEre_CAF1 52215 118 + 1 AAAACAGCAACAAUAAUGUGUUAAUGUGUGCAAACACACACAAAUGUAUGUAUGUAUAUACAUACAUGCGAAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGAAGAAAUU ...............(((((((..((((((......))))))..(((((((((((....))))))))))).....-.((....))(((.....)))..))))))).............. ( -33.20) >DroYak_CAF1 43808 115 + 1 AAAACAGCAAAAAUAAUGUGUUAAUGUGUGCAAACACACACAC---UGUGUAUGUAUAGACAUACAUGUAAAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAUAUU ...............(((((((..((((((......)))))).---.((((((((....))))))))........-.((....))(((.....)))..))))))).............. ( -28.20) >consensus AAAACAGCAACAAUAAUGUGUUAAUGUGUGCAAACACACACAAAUGCAUC____UGCAUACAUACA__UAGAGUA_AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUU ...............(((((((..((((((......)))))).(((((......)))))..................((....))(((.....)))..))))))).............. (-22.28 = -22.12 + -0.16)

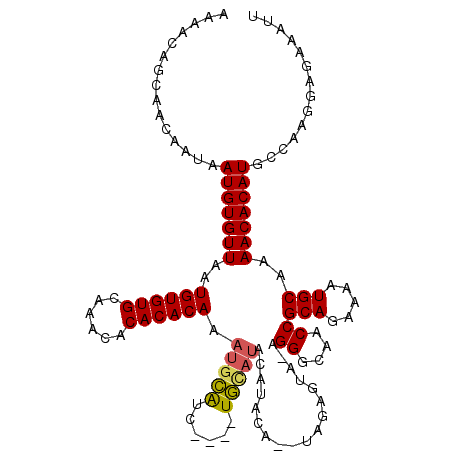
| Location | 11,784,079 – 11,784,190 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -18.15 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11784079 111 - 22407834 AAUUUCUCCUUGGCAUGUGUUUUGCAUUUUCUGCGGUUGCCCUAUACUCUA----CAUGUAUGCA----GAUGCAUUUGUGUGUGUUUGCACACAUUAACACAUUAUUGUUGCUGUUUU ...........((((((((((.((((...(((((((....)).((((....----...)))))))----))))))..(((((((....)))))))..)))))).......))))..... ( -27.81) >DroSec_CAF1 41846 112 - 1 AAUUUCUCGUUGGCAUGUGUUUUGCAUUUUCUGCGGUUGCCCU-UACUCUA--UGUAUGUAUGCA----GAUGCAUUUGUGUGUGUUUGCACACAUUAACACAUUAUUGUUGCUGUUUU ...........((((((((((.((((...((((((..(((...-(((....--.))).)))))))----))))))..(((((((....)))))))..)))))).......))))..... ( -27.71) >DroSim_CAF1 40950 112 - 1 AAUUUCUCCUUGGCAUGUGUUUUGCAUUUUCUGCGGUUGCCCU-UACUCUA--UGUAUGUAUGCA----GAUGCAUUUGUGUGUGUUUGCACACAUUAACACAUUAUUGUUGCUGUUUU ...........((((((((((.((((...((((((..(((...-(((....--.))).)))))))----))))))..(((((((....)))))))..)))))).......))))..... ( -27.71) >DroEre_CAF1 52215 118 - 1 AAUUUCUUCUUGGCAUGUGUUUUGCAUUUUCUGCGGUUGCCCU-UACUUCGCAUGUAUGUAUAUACAUACAUACAUUUGUGUGUGUUUGCACACAUUAACACAUUAUUGUUGCUGUUUU ...........((((((((((...........((((.......-....))))((((((((((....)))))))))).(((((((....)))))))..)))))).......))))..... ( -27.91) >DroYak_CAF1 43808 115 - 1 AAUAUCUCCUUGGCAUGUGUUUUGCAUUUUCUGCGGUUGCCCU-UACUUUACAUGUAUGUCUAUACAUACACA---GUGUGUGUGUUUGCACACAUUAACACAUUAUUUUUGCUGUUUU ...........(((((((((((((((.....))))).......-.........(((((((....))))))).(---((((((((....))))))))))))))).......))))..... ( -25.61) >consensus AAUUUCUCCUUGGCAUGUGUUUUGCAUUUUCUGCGGUUGCCCU_UACUCUA__UGUAUGUAUGCA____GAUGCAUUUGUGUGUGUUUGCACACAUUAACACAUUAUUGUUGCUGUUUU ...........((((((((((.(((((....((.(((........))).)).....)))))................(((((((....)))))))..)))))).......))))..... (-18.15 = -18.03 + -0.12)

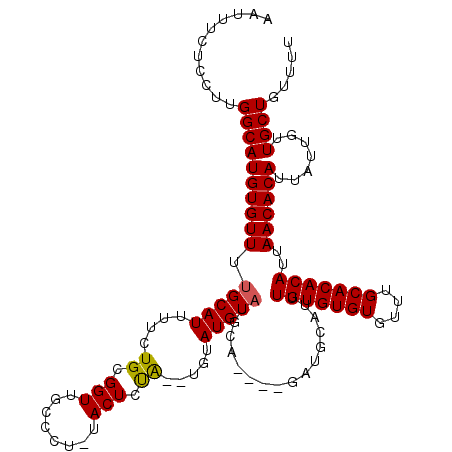
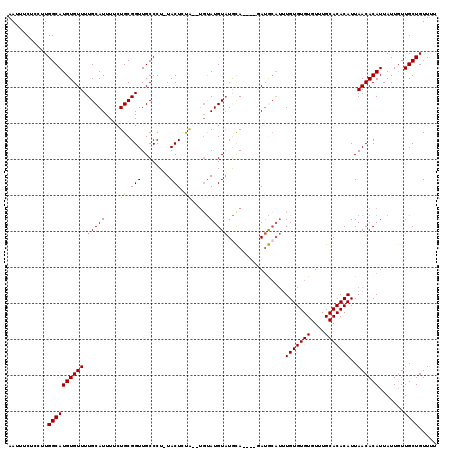
| Location | 11,784,118 – 11,784,226 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.56 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -18.88 |
| Energy contribution | -20.56 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11784118 108 + 22407834 ACAAAUGCAUC----UGCAUACAUG----UAGAGUAUAGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACA .....((((((----(((((((...----....)))).((....)))))))...))))...(((((.((..(((....)))(((((......).)))).))))))).......... ( -30.40) >DroSec_CAF1 41885 109 + 1 ACAAAUGCAUC----UGCAUACAUACA--UAGAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAACGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACA .....((((((----(((.....(((.--....)))-.((....)))))))...))))...(((((.((...(((...)))(((((......).)))).))))))).......... ( -26.30) >DroSim_CAF1 40989 109 + 1 ACAAAUGCAUC----UGCAUACAUACA--UAGAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACA .....((((((----(((.....(((.--....)))-.((....)))))))...))))...(((((.((..(((....)))(((((......).)))).))))))).......... ( -29.70) >DroEre_CAF1 52254 114 + 1 ACAAAUGUAUGUAUGUAUAUACAUACAUGCGAAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGAAGAAAUUCGCUCCAUUGAAGAGAGUA-GAUGUGGAAAAGGACA .....(((((((((((....))))))))))).....-.((....))(((.....)))....(((((.....(((....)))(((((......).)))).-.))))).......... ( -31.10) >DroYak_CAF1 43847 112 + 1 ACAC---UGUGUAUGUAUAGACAUACAUGUAAAGUA-AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAUAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACA ....---.((((((((....))))))))........-.((....))(((.....)))....(((((.((..(((....)))(((((......).)))).))))))).......... ( -27.10) >consensus ACAAAUGCAUC____UGCAUACAUACA__UAGAGUA_AGGGCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACA ....(((((......)))))..................((....))(((.....)))....(((((.((..(((....)))(((((......).)))).))))))).......... (-18.88 = -20.56 + 1.68)

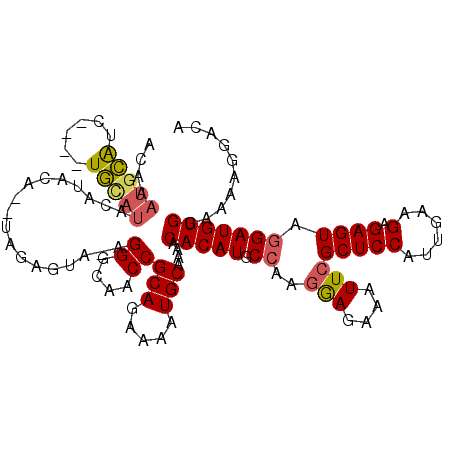
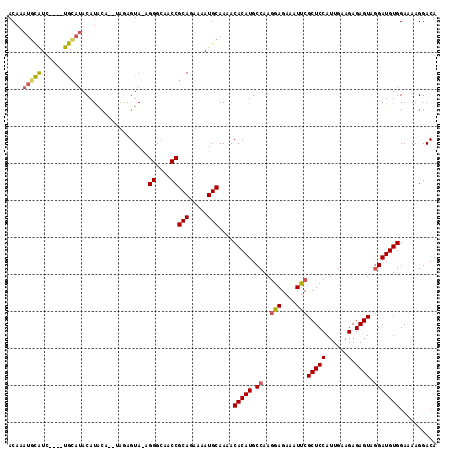
| Location | 11,784,150 – 11,784,266 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 98.28 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -28.62 |
| Energy contribution | -30.46 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11784150 116 + 22407834 GCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACACACCGGAUGUGGCACUGCAUCCGAAGGAUGAUGAUGCAAG (((.(((((.....)))....(((((.((..(((....)))(((((......).)))).))))))).....))......(((((((......)))))))...........)))... ( -32.70) >DroSec_CAF1 41918 116 + 1 GCAACCGCAGAAAAUGCAAAACACAUGCCAACGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACACACCGGAUGUGGCACUGCAUCCGAAGGAUGAUGAUGCAAG ((....(((.....)))....(((((.((............(((((......).)))).((.((((........)))))))))))))))..(((((((....).....)))))).. ( -29.70) >DroSim_CAF1 41022 116 + 1 GCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACACACCGGAUGUGGCACUGCAUCCGAAGGAUGAUGAUGCAAG (((.(((((.....)))....(((((.((..(((....)))(((((......).)))).))))))).....))......(((((((......)))))))...........)))... ( -32.70) >DroEre_CAF1 52293 115 + 1 GCAACCGCAGAAAAUGCAAAACACAUGCCAAGAAGAAAUUCGCUCCAUUGAAGAGAGUA-GAUGUGGAAAAGGACACAGCGGAUGUGGCACUGCAUCCGAAGGAUGAUGAUGCAAG (((.(((((.....)))....(((((.....(((....)))(((((......).)))).-.))))).....)).(((..(((((((......)))))))...).))....)))... ( -30.60) >DroYak_CAF1 43883 116 + 1 GCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAUAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACACACCGGAUGUGGCACUGCAUCCGAAGGAUGAUGAUGCAAG ((....(((.....)))....(((((.((..((((.......)))).............((.((((........)))))))))))))))..(((((((....).....)))))).. ( -31.70) >consensus GCAACCGCAGAAAAUGCAAAACACAUGCCAAGGAGAAAUUCGCUCCAUUGAAGAGAGUAGGAUGUGGAAAAGGACACACCGGAUGUGGCACUGCAUCCGAAGGAUGAUGAUGCAAG (((.(((((.....)))....(((((.((..(((....)))(((((......).)))).))))))).....))......(((((((......)))))))...........)))... (-28.62 = -30.46 + 1.84)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:17 2006