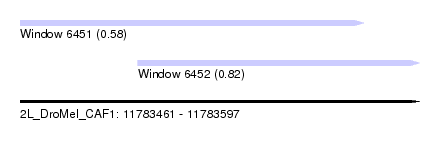
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,783,461 – 11,783,597 |
| Length | 136 |
| Max. P | 0.816406 |

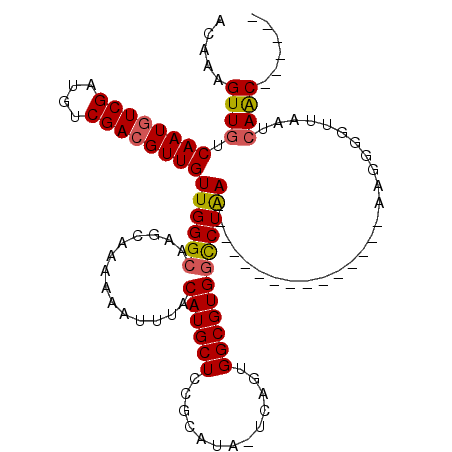
| Location | 11,783,461 – 11,783,578 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.36 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -24.42 |
| Energy contribution | -25.73 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11783461 117 + 22407834 CGGCAACAGCAGAAAGAAAUUACACGCAAGCAACAAGACAACAAAGUUGUCAAUGUCGAUG---UCGAAGUUGUUGGGCAAGCAAAAAAUUUACAUGCUCCGCAUGCUCAGUGGCGUGGC .(....)...............(((((.((((((..((((((...))))))....(((...---.))).))))))(((((.((..................)).)))))....))))).. ( -31.67) >DroSec_CAF1 41198 116 + 1 CGGCAACAGCAGAAAGAAAUUACACGCAAGCAACAAGACAACAAAGUUGUCAAUGUCGAUG---UCGACGUUGUUGGGCAAGCAAAAAAUGUACAUGCUCAGCAUA-UCAGUGGCGUGGC .(....)...............(((((..((.....((((((...))))))(((((((...---.)))))))((((((((.(((.....)))...))))))))...-...)).))))).. ( -35.00) >DroSim_CAF1 40292 116 + 1 CGGCAACAGCAGAAAGAAAUUACACGCAAGCAACAAGACAACAAAGUUGUCAAUGUCGAUG---UCGACGUUGUUGGGCAAGCAAAAAAUUUACAUGCUCCGCAUA-UCAGUGGCGUGGC .(....).((.(.((....)).)..((.((((((..((((((...))))))...((((...---.))))))))))..))..))..........(((((..(.....-...)..))))).. ( -32.50) >DroEre_CAF1 51579 116 + 1 CGGCAACAGCAGAAAGAAAUUACACGCAAGCAACAAGACAACAAAGUUGUCAAUGUCGAUG---UCGACGUUGUUGGGCGAGCAAAGAAUUUACAUGCUCCGCAUA-UCAGGGGCGUGGC .(....).((.(.((....)).).(((.((((((..((((((...))))))...((((...---.))))))))))..))).))..........((((((((.....-...)))))))).. ( -39.80) >DroYak_CAF1 43076 116 + 1 GGGCAACAGCAGAAAGAAAUUACACGCAAGCAACAAGACAACAAAGUUGUCAAUGUCGAUG---UCGACGUUGUUGGGCAAACAAAAAAUUUACAUGCUCCUCAUA-UCAGGGGCGUGUU .(....)..................((.((((((..((((((...))))))...((((...---.))))))))))..)).............((((((((((....-..)))))))))). ( -36.10) >DroAna_CAF1 42338 97 + 1 CGGCAACAGCAGAAAGAAAUUACAUGCAAACAGCAAGACAACAAAGUUGUCAAUGUCGAUGUUGUUGACGUUGUUUGGCUUGCAACAAAUUUCCACU----------------------- ..(((..(((.(.((....)).)...((((((((..((((((...))))))...((((((...))))))))))))))))))))..............----------------------- ( -25.60) >consensus CGGCAACAGCAGAAAGAAAUUACACGCAAGCAACAAGACAACAAAGUUGUCAAUGUCGAUG___UCGACGUUGUUGGGCAAGCAAAAAAUUUACAUGCUCCGCAUA_UCAGUGGCGUGGC .(....)...............(((((.((((((..((((((...))))))...(((((.....)))))))))))(((((...............))))).............))))).. (-24.42 = -25.73 + 1.31)

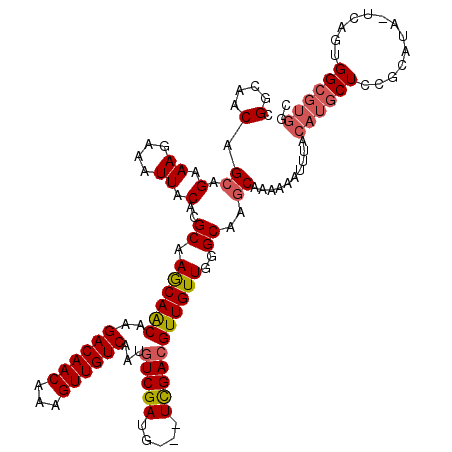
| Location | 11,783,501 – 11,783,597 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -30.72 |
| Consensus MFE | -18.61 |
| Energy contribution | -18.65 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11783501 96 + 22407834 ACAAAGUUGUCAAUGUCGAUGUCGAAGUUGUUGGGCAAGCAAAAAAUUUACAUGCUCCGCAUGCUCAGUGGCGUGGCCUAA---------------AAGGGAUUAAUAAAC------ ..........((((.(((....))).))))((((((.((((...........))))...((((((....))))))))))))---------------...............------ ( -20.50) >DroSec_CAF1 41238 95 + 1 ACAAAGUUGUCAAUGUCGAUGUCGACGUUGUUGGGCAAGCAAAAAAUGUACAUGCUCAGCAUA-UCAGUGGCGUGGCCUAA---------------AGGGGGUUAAUCAAC------ ........((((((((((....))))))(((((((((.(((.....)))...)))))))))..-....)))).((((((..---------------...))))))......------ ( -26.70) >DroSim_CAF1 40332 95 + 1 ACAAAGUUGUCAAUGUCGAUGUCGACGUUGUUGGGCAAGCAAAAAAUUUACAUGCUCCGCAUA-UCAGUGGCGUGGCCUAA---------------AAGGGGUUAAUCAAC------ .....((((.((((((((....))))))))((((((.((((...........))))((((...-...))))....))))))---------------...........))))------ ( -24.80) >DroEre_CAF1 51619 110 + 1 ACAAAGUUGUCAAUGUCGAUGUCGACGUUGUUGGGCGAGCAAAGAAUUUACAUGCUCCGCAUA-UCAGGGGCGUGGCCUGAUGAAUUCCCCUUUUAAAGGGGCUCAUCAGC------ .....(((..((((((((....))))))))...)))..............((((((((.....-...))))))))..(((((((...(((((.....))))).))))))).------ ( -41.70) >DroYak_CAF1 43116 116 + 1 ACAAAGUUGUCAAUGUCGAUGUCGACGUUGUUGGGCAAACAAAAAAUUUACAUGCUCCUCAUA-UCAGGGGCGUGUUCUGAAGAAUUUCCCUUUCAAAGGGGCUUAACAACUUUUAA ..((((((((((((((((....)))))))).(((((.............((((((((((....-..))))))))))............((((.....)))))))))))))))))... ( -39.90) >consensus ACAAAGUUGUCAAUGUCGAUGUCGACGUUGUUGGGCAAGCAAAAAAUUUACAUGCUCCGCAUA_UCAGUGGCGUGGCCUAA_______________AAGGGGUUAAUCAAC______ .....((((.((((((((....))))))))((((((..............((((((.............))))))))))))..........................))))...... (-18.61 = -18.65 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:12 2006