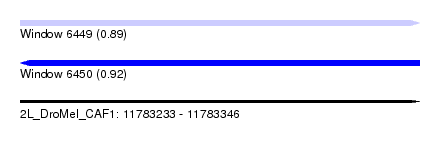
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,783,233 – 11,783,346 |
| Length | 113 |
| Max. P | 0.920503 |

| Location | 11,783,233 – 11,783,346 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -34.74 |
| Energy contribution | -34.78 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11783233 113 + 22407834 CUCCUCGACUUUUCCCCAGGCGACUUGCCUUUG------CCUUUGCGUUUGACACUUUUCCACAGUAACGGAAGUGGGACUGCUGGGGAACUAUCAAAGUGCACCUAGGCGAGGGGGAC ...........((((((((((.....))))(((------(((.(((((((((.(.((..(((((((.((....))...)))).)))..)).).))))).))))...)))))))))))). ( -44.90) >DroSec_CAF1 40973 110 + 1 CUCCUCGACUUUUCCCCAGGCGACUUGCCUUUG------CCUUUGCGUUUGACACUUUUCCACAGUAACGGAAGUGGGGCUGCUGGGGAACUAUCAAAGUGCACCUAGAC---GGGGAC ............(((((((((((.......)))------))).(((((((((.(.((..(((((((.((....))...)))).)))..)).).))))).)))).......---))))). ( -38.00) >DroSim_CAF1 40067 110 + 1 CUCCUCGACUUUUCCCCAGGCGACUUGCCUUUG------CCUUUGCGUUUGACACUUUUCCACAGUAACGGAAGUGGGGCUGCUGGGGAACUAUCAAAGUGCACCUAGAC---GGGGAC ............(((((((((((.......)))------))).(((((((((.(.((..(((((((.((....))...)))).)))..)).).))))).)))).......---))))). ( -38.00) >DroEre_CAF1 51343 109 + 1 CUCCUCGACUUUUCCCCAGGCGACUUGCCUUUG------CGUUUGCGUUUGACACUUUUCCGCAGUAACGGAAGUGG-GCUGCUGGGGAACUAUCAAAGUGCACCUAGAC---GGGGAC ............(((((((((.....))))...------.((((((((((((.(.((..(((((((.((....))..-))))).))..)).).))))).))))....)))---))))). ( -38.70) >DroYak_CAF1 42856 115 + 1 CUCCUCGACUUUUCCCCAGGCGACUUGCCUUUGCGAGUGCGUUUGCGUUUGACACUUUUCCGCAGAAACGGAAGUGG-GCUGCUGGGGAACUAUCAUAGUGCACCUAGAC---GGGGAC .((((((....((((((((((.((((((....))))))))(((..((.....)....(((((......))))))..)-))..))))))))...((.(((.....))))))---))))). ( -41.30) >consensus CUCCUCGACUUUUCCCCAGGCGACUUGCCUUUG______CCUUUGCGUUUGACACUUUUCCACAGUAACGGAAGUGGGGCUGCUGGGGAACUAUCAAAGUGCACCUAGAC___GGGGAC ............(((((((((.....)))).............(((((((((.(.((..(((((((.((....))...)))).)))..)).).))))).))))..........))))). (-34.74 = -34.78 + 0.04)

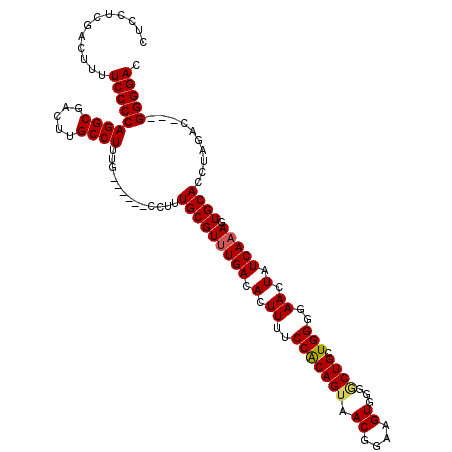
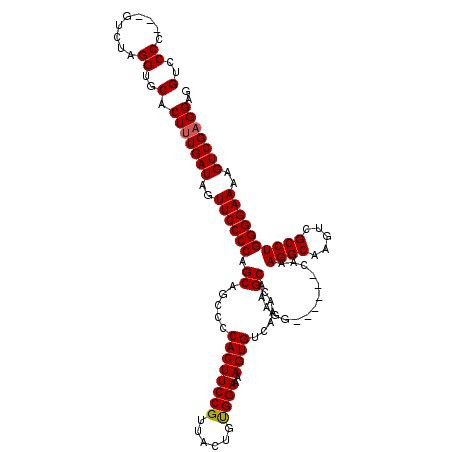
| Location | 11,783,233 – 11,783,346 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -38.56 |
| Consensus MFE | -33.98 |
| Energy contribution | -33.94 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920503 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11783233 113 - 22407834 GUCCCCCUCGCCUAGGUGCACUUUGAUAGUUCCCCAGCAGUCCCACUUCCGUUACUGUGGAAAAGUGUCAAACGCAAAGG------CAAAGGCAAGUCGCCUGGGGAAAAGUCGAGGAG .(((((...((((...(((..(((((((.((..(((.((((..(......)..)))))))..)).))))))).))).)))------)..((((.....)))))))))............ ( -38.80) >DroSec_CAF1 40973 110 - 1 GUCCCC---GUCUAGGUGCACUUUGAUAGUUCCCCAGCAGCCCCACUUCCGUUACUGUGGAAAAGUGUCAAACGCAAAGG------CAAAGGCAAGUCGCCUGGGGAAAAGUCGAGGAG (..((.---.....))..).(((((((..((((((....(((((((..........))))....(((.....)))...))------)..((((.....))))))))))..))))))).. ( -38.90) >DroSim_CAF1 40067 110 - 1 GUCCCC---GUCUAGGUGCACUUUGAUAGUUCCCCAGCAGCCCCACUUCCGUUACUGUGGAAAAGUGUCAAACGCAAAGG------CAAAGGCAAGUCGCCUGGGGAAAAGUCGAGGAG (..((.---.....))..).(((((((..((((((....(((((((..........))))....(((.....)))...))------)..((((.....))))))))))..))))))).. ( -38.90) >DroEre_CAF1 51343 109 - 1 GUCCCC---GUCUAGGUGCACUUUGAUAGUUCCCCAGCAGC-CCACUUCCGUUACUGCGGAAAAGUGUCAAACGCAAACG------CAAAGGCAAGUCGCCUGGGGAAAAGUCGAGGAG (..((.---.....))..).(((((((..((((((.((...-....((((((....))))))..(((.....)))....)------)..((((.....))))))))))..))))))).. ( -39.20) >DroYak_CAF1 42856 115 - 1 GUCCCC---GUCUAGGUGCACUAUGAUAGUUCCCCAGCAGC-CCACUUCCGUUUCUGCGGAAAAGUGUCAAACGCAAACGCACUCGCAAAGGCAAGUCGCCUGGGGAAAAGUCGAGGAG (..((.---.....))..).((.((((..((((((.((...-.(((((((((....)))))..))))......((....))....))..((((.....))))))))))..)))).)).. ( -37.00) >consensus GUCCCC___GUCUAGGUGCACUUUGAUAGUUCCCCAGCAGCCCCACUUCCGUUACUGUGGAAAAGUGUCAAACGCAAAGG______CAAAGGCAAGUCGCCUGGGGAAAAGUCGAGGAG (..((.........))..).(((((((..((((((.((.....((((((((......))))..))))......))..............((((.....))))))))))..))))))).. (-33.98 = -33.94 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:10 2006