| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,777,020 – 11,777,134 |
| Length | 114 |
| Max. P | 0.955573 |

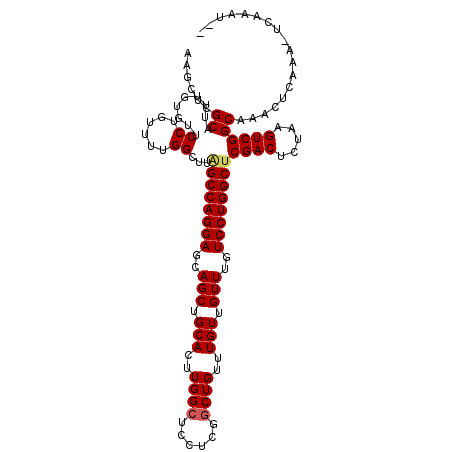
| Location | 11,777,020 – 11,777,134 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -27.78 |
| Energy contribution | -28.06 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11777020 114 + 22407834 AAGCUCGCAUUUGUGUUCCUGUU-UGGAUUAGCCAGGAGCAGCUGCACUUGGCUCCUCGACUGUUUGUUGUUUGUCCUGGCUCGACUCUAAGUCGGCAAACUCAAAUUCAAAUUC ......(.(((((.(((((..((-(((((((((((((((((((.(((.((((....)))).)))..)))))...)))))))).)).))))))..))..))).))))).)...... ( -33.30) >DroSec_CAF1 34953 113 + 1 AAGCUCGCAUUUGUGUUCCUGUUUUGGCUUAGCCAGGAGCAGCUGCACUUGGCUCCUCGGCUGUUUGUUGUUUGUCCUGGCUCGACUCUAAGUCGGCAAACUCAAACUCAAAU-- .((((.((....((((..((((((((((...)))).))))))..))))...)).....))))(((((..(((((.....((.((((.....)))))).....))))).)))))-- ( -35.40) >DroSim_CAF1 34043 113 + 1 AAGCUCGCAUUUGUGUUCCUGUUUUGGCUUAGCCAGGAGCAGCUGCACUUGGCUCCUCGGCUGUUUGUUGUUUGUCCUGGCUCGACUCUAAGUCGGCAAACUCAAACUCAAAU-- .((((.((....((((..((((((((((...)))).))))))..))))...)).....))))(((((..(((((.....((.((((.....)))))).....))))).)))))-- ( -35.40) >DroEre_CAF1 45448 107 + 1 AAGCCUGCAUUGCUGCUCCUGUUUUGGCUUUGCCAGGAGCAGCUGCACUUGGCUCCUCUGCUGUUUGUUGUUUGUCCUGGCUCGACUCUAAGUCGGCAAAUUC------AAAU-- .((((((((..(((((((((.....(((...))))))))))))))))...))))........(((((..(((((.((.((((........))))))))))).)------))))-- ( -37.20) >DroYak_CAF1 36525 107 + 1 AAACUUGCAUUUGUGUUCCUAUUUCGGCUUGGCCAGGAGCAGCUGCACUUGGCUCCUUUGCUGUUUGUUGUUUGUCCUGGCUCGACUCUAAGUCGGCAAAUUC------AAAU-- ....(((.((((((................((((((((..(((.(((..((((......))))..))).)))..))))))))((((.....)))))))))).)------))..-- ( -33.10) >consensus AAGCUCGCAUUUGUGUUCCUGUUUUGGCUUAGCCAGGAGCAGCUGCACUUGGCUCCUCGGCUGUUUGUUGUUUGUCCUGGCUCGACUCUAAGUCGGCAAACUCAAA_UCAAAU__ ......((.........((......))...((((((((..(((.(((..((((......))))..))).)))..))))))))((((.....)))))).................. (-27.78 = -28.06 + 0.28)


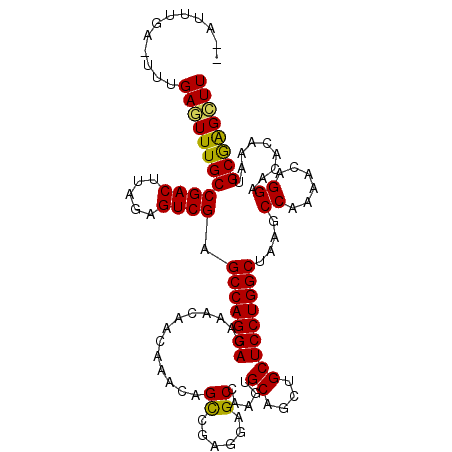
| Location | 11,777,020 – 11,777,134 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -26.78 |
| Energy contribution | -26.18 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11777020 114 - 22407834 GAAUUUGAAUUUGAGUUUGCCGACUUAGAGUCGAGCCAGGACAAACAACAAACAGUCGAGGAGCCAAGUGCAGCUGCUCCUGGCUAAUCCA-AACAGGAACACAAAUGCGAGCUU ............((((((((((((.....))))(((((((((............)))(((.(((........))).)))))))))..(((.-....)))........)))))))) ( -34.00) >DroSec_CAF1 34953 113 - 1 --AUUUGAGUUUGAGUUUGCCGACUUAGAGUCGAGCCAGGACAAACAACAAACAGCCGAGGAGCCAAGUGCAGCUGCUCCUGGCUAAGCCAAAACAGGAACACAAAUGCGAGCUU --....(((((((.(((((.((((.....))))..((.(......).......((((.((((((..((.....))))))))))))...........))....))))).))))))) ( -31.00) >DroSim_CAF1 34043 113 - 1 --AUUUGAGUUUGAGUUUGCCGACUUAGAGUCGAGCCAGGACAAACAACAAACAGCCGAGGAGCCAAGUGCAGCUGCUCCUGGCUAAGCCAAAACAGGAACACAAAUGCGAGCUU --....(((((((.(((((.((((.....))))..((.(......).......((((.((((((..((.....))))))))))))...........))....))))).))))))) ( -31.00) >DroEre_CAF1 45448 107 - 1 --AUUU------GAAUUUGCCGACUUAGAGUCGAGCCAGGACAAACAACAAACAGCAGAGGAGCCAAGUGCAGCUGCUCCUGGCAAAGCCAAAACAGGAGCAGCAAUGCAGGCUU --.(((------(..((.((((((.....)))).)).))..))))...............(((((...((((((((((((((............))))))))))..))))))))) ( -38.10) >DroYak_CAF1 36525 107 - 1 --AUUU------GAAUUUGCCGACUUAGAGUCGAGCCAGGACAAACAACAAACAGCAAAGGAGCCAAGUGCAGCUGCUCCUGGCCAAGCCGAAAUAGGAACACAAAUGCAAGUUU --....------((((((((((((.....)))).(((((((.............((......)).....((....)))))))))....((......)).........)))))))) ( -26.40) >consensus __AUUUGA_UUUGAGUUUGCCGACUUAGAGUCGAGCCAGGACAAACAACAAACAGCCGAGGAGCCAAGUGCAGCUGCUCCUGGCUAAGCCAAAACAGGAACACAAAUGCGAGCUU ............((((((((((((.....)))).(((((((.............((......)).....((....)))))))))....((......)).........)))))))) (-26.78 = -26.18 + -0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:05 2006