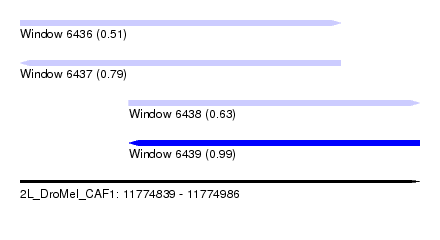
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,774,839 – 11,774,986 |
| Length | 147 |
| Max. P | 0.991675 |

| Location | 11,774,839 – 11,774,957 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -32.44 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.85 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
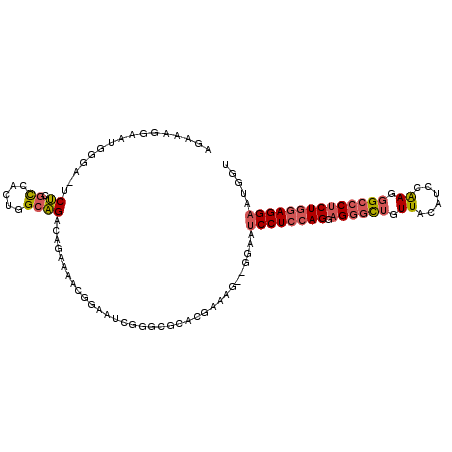
>2L_DroMel_CAF1 11774839 118 + 22407834 UGUUGUCCAAUUCAGUAAUCCAAGGCAUCAGGAACAGCCGACCAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCU--CUUUCGUGCGCCCGAUUCAGUUUUCUGU ............(((.((..(...(.(((.((....(((((....(((((((((((((..(((....)))..))))).))))))))....--...))).)).)).))).).)..))))). ( -33.50) >DroSec_CAF1 32865 118 + 1 UGUUGUCCAAUUCAGUAAUCCAAGGCAUCAGGAACAGCCGACCAUUCCUCCAGUGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCC--CUUUCGUGCGCCCGAUUCCGUUUUCUGU ............(((........((.(((.((....(((((....((((((((.((((..(((....)))..))))..))))))))....--...))).)).)).))).)).....))). ( -33.82) >DroSim_CAF1 31966 118 + 1 UGUUGUCCAAUUCAGUAAUCCAAGGCAUCAGGAACAGCCGACCAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCC--CUUUCGUGCGCCCGAUUCCGUUUUCUGU ............(((........((.(((.((....(((((....(((((((((((((..(((....)))..))))).))))))))....--...))).)).)).))).)).....))). ( -36.22) >DroEre_CAF1 43219 113 + 1 UGUUGUCCAAUUCAGUAAUCCAAGGCAUCAGGAACAGCCGACGAUUCCUCCAGAGGGCCCUUGGAAGUAACAGCCCUCCUGGAGGAUUCC--CUC-----CUCCCGAUUCCGCUUUCUGU ............(((........((.(((.(((..((.....((.(((((((((((((..(((....)))..))))).)))))))).)).--)).-----.))).))).)).....))). ( -33.52) >DroYak_CAF1 34332 99 + 1 UGUUGUCCAAUUCAGUAAUCCAAGGCAUCAGGAACAGCCGACAAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAU---------------------UCCGUUUUCUGU ((.((((................)))).))...((((..(((...(((((((((((((..(((....)))..))))).)))))))).---------------------...)))..)))) ( -29.99) >DroAna_CAF1 35147 116 + 1 GGUUGUCCAAUUCACUAAACCAAGGCAUCAAAAUUAGCCUUGAGAACCUCUAGAGUGCCAUCGACCAUAACCAAGCUCCUGCAGGACUCGAGCUUUUGU---UUUGUUUCAGCUUU-UAU (((((....((((.......((((((..........))))))((.....)).)))).....)))))......(((((...(((((((..........))---)))))...))))).-... ( -27.60) >consensus UGUUGUCCAAUUCAGUAAUCCAAGGCAUCAGGAACAGCCGACCAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCC__CUUUCGU_CGCCCGAUUCCGUUUUCUGU .......................(((..........)))......(((((((((((((..(((....)))..))))).)))))))).................................. (-22.26 = -22.85 + 0.59)

| Location | 11,774,839 – 11,774,957 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.75 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -26.88 |
| Energy contribution | -27.80 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11774839 118 - 22407834 ACAGAAAACUGAAUCGGGCGCACGAAAG--AGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUGGUCGGCUGUUCCUGAUGCCUUGGAUUACUGAAUUGGACAACA .(((.((.(..(((((((.((((....)--.((.((((((((.((((((.((......)).))))))))))))))....))...))).))))))...)..).)).)))............ ( -41.70) >DroSec_CAF1 32865 118 - 1 ACAGAAAACGGAAUCGGGCGCACGAAAG--GGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCACUGGAGGAAUGGUCGGCUGUUCCUGAUGCCUUGGAUUACUGAAUUGGACAACA ........(((((((((((((((....)--((((((((((((..(((((.((......)).))))).))))))))..((....)).)))))).)))))..)))).)))............ ( -41.10) >DroSim_CAF1 31966 118 - 1 ACAGAAAACGGAAUCGGGCGCACGAAAG--GGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUGGUCGGCUGUUCCUGAUGCCUUGGAUUACUGAAUUGGACAACA ........(((((((((((((((....)--((((((((((((.((((((.((......)).))))))))))))))..((....)).)))))).)))))..)))).)))............ ( -42.60) >DroEre_CAF1 43219 113 - 1 ACAGAAAGCGGAAUCGGGAG-----GAG--GGAAUCCUCCAGGAGGGCUGUUACUUCCAAGGGCCCUCUGGAGGAAUCGUCGGCUGUUCCUGAUGCCUUGGAUUACUGAAUUGGACAACA .(((.((..((.((((((((-----.((--.((.((((((((.((((((.((......)).)))))))))))))).)).....)).)))))))).)).....)).)))............ ( -43.70) >DroYak_CAF1 34332 99 - 1 ACAGAAAACGGA---------------------AUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUUGUCGGCUGUUCCUGAUGCCUUGGAUUACUGAAUUGGACAACA ............---------------------.((((((((.((((((.((......)).)))))))))))))).((((((((..........)))(..(....)..)....))))).. ( -35.40) >DroAna_CAF1 35147 116 - 1 AUA-AAAGCUGAAACAAA---ACAAAAGCUCGAGUCCUGCAGGAGCUUGGUUAUGGUCGAUGGCACUCUAGAGGUUCUCAAGGCUAAUUUUGAUGCCUUGGUUUAGUGAAUUGGACAACC ...-..........((.(---((..((((((..(.....)..)))))).))).))(((.(...((((...(((((..((((((....)))))).))))).....))))...).))).... ( -25.50) >consensus ACAGAAAACGGAAUCGGGCG_ACGAAAG__GGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUGGUCGGCUGUUCCUGAUGCCUUGGAUUACUGAAUUGGACAACA .(((..............................((((((((.((((((.((......)).))))))))))))))......(((..........)))........)))............ (-26.88 = -27.80 + 0.92)


| Location | 11,774,879 – 11,774,986 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -30.48 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629049 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11774879 107 + 22407834 ACCAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCU--CUUUCGUGCGCCCGAUUCAGUUUUCUGUCUGCCAGGGGCGAGA-UCCCAUUCCUUUCU .....(((((((((((((..(((....)))..))))).))))))))....--...((...(((((....(((........)))....))))).))-.............. ( -36.10) >DroSec_CAF1 32905 107 + 1 ACCAUUCCUCCAGUGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCC--CUUUCGUGCGCCCGAUUCCGUUUUCUGUCUGCCAGUGGCGAGA-UCCCAUUCCUUUCU .....((((((((.((((..(((....)))..))))..))))))))....--.............(((..((((..(((.....))).))))..)-))............ ( -29.90) >DroSim_CAF1 32006 107 + 1 ACCAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCC--CUUUCGUGCGCCCGAUUCCGUUUUCUGUCUGCCAGUGGCGAGU-UCCCAUUCCUUUCU .....(((((((((((((..(((....)))..))))).))))))))....--........((((((....))....(((.....))).))))...-.............. ( -29.80) >DroEre_CAF1 43259 101 + 1 ACGAUUCCUCCAGAGGGCCCUUGGAAGUAACAGCCCUCCUGGAGGAUUCC--CUC-----CUCCCGAUUCCGCUUUCUGUCUGCCAG-GACGAGGUU-CCCAUUCCUGCU ..((.(((((((((((((..(((....)))..))))).)))))))).))(--(((-----.(((.......((.....))......)-)).))))..-............ ( -33.92) >DroAna_CAF1 35187 98 + 1 UGAGAACCUCUAGAGUGCCAUCGACCAUAACCAAGCUCCUGCAGGACUCGAGCUUUUGU---UUUGUUUCAGCUUU-UAUCCUCCAGCCGGAGGA-UACCCCU------- (((((.......((((.((...(........)..((....)).))))))((((....))---))..))))).....-((((((((....))))))-)).....------- ( -22.70) >consensus ACCAUUCCUCCAGAGGGCCCUUGGAUGUAACAGCCCUCCUGGAGGAUUCC__CUUUCGUGCGCCCGAUUCCGUUUUCUGUCUGCCAGUGGCGAGA_UCCCAUUCCUUUCU .....(((((((((((((..(((....)))..))))).)))))))).................................(((((.....)).)))............... (-18.94 = -19.30 + 0.36)



| Location | 11,774,879 – 11,774,986 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -21.46 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11774879 107 - 22407834 AGAAAGGAAUGGGA-UCUCGCCCCUGGCAGACAGAAAACUGAAUCGGGCGCACGAAAG--AGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUGGU ..............-((((((((....(((........)))....)))))..(....)--))).((((((((.((((((.((......)).))))))))))))))..... ( -40.40) >DroSec_CAF1 32905 107 - 1 AGAAAGGAAUGGGA-UCUCGCCACUGGCAGACAGAAAACGGAAUCGGGCGCACGAAAG--GGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCACUGGAGGAAUGGU ..............-(((((((.(((.....)))....((....))))))..(....)--))).((((((((..(((((.((......)).))))).))))))))..... ( -36.80) >DroSim_CAF1 32006 107 - 1 AGAAAGGAAUGGGA-ACUCGCCACUGGCAGACAGAAAACGGAAUCGGGCGCACGAAAG--GGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUGGU ..............-...((((.(((.....)))....((....))))))..(....)--....((((((((.((((((.((......)).))))))))))))))..... ( -37.80) >DroEre_CAF1 43259 101 - 1 AGCAGGAAUGGG-AACCUCGUC-CUGGCAGACAGAAAGCGGAAUCGGGAG-----GAG--GGAAUCCUCCAGGAGGGCUGUUACUUCCAAGGGCCCUCUGGAGGAAUCGU ............-..((((.((-((((....(.......)...)))))).-----)))--)((.((((((((.((((((.((......)).)))))))))))))).)).. ( -43.70) >DroAna_CAF1 35187 98 - 1 -------AGGGGUA-UCCUCCGGCUGGAGGAUA-AAAGCUGAAACAAA---ACAAAAGCUCGAGUCCUGCAGGAGCUUGGUUAUGGUCGAUGGCACUCUAGAGGUUCUCA -------.((((..-.((((...(((.(((((.-..((((........---.....))))...))))).)))(((....(((((.....))))).)))..)))).)))). ( -27.62) >consensus AGAAAGGAAUGGGA_UCUCGCCACUGGCAGACAGAAAACGGAAUCGGGCGCACGAAAG__GGAAUCCUCCAGGAGGGCUGUUACAUCCAAGGGCCCUCUGGAGGAAUGGU ................((.((.....))))..................................((((((((.((((((.((......)).))))))))))))))..... (-21.46 = -22.26 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:59 2006