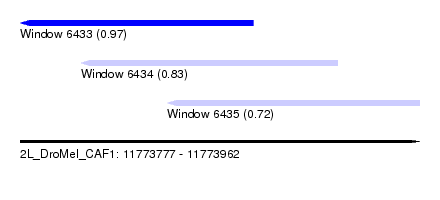
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,773,777 – 11,773,962 |
| Length | 185 |
| Max. P | 0.974451 |

| Location | 11,773,777 – 11,773,885 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -15.42 |
| Energy contribution | -15.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
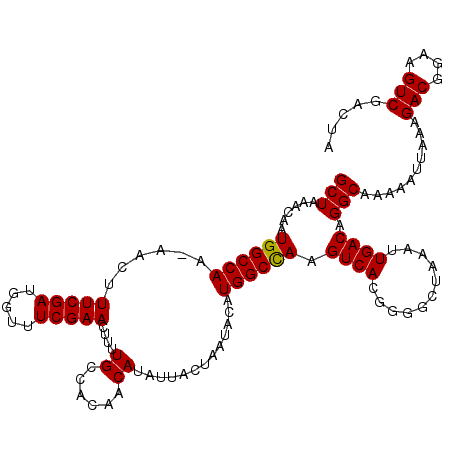
>2L_DroMel_CAF1 11773777 108 - 22407834 GCUAAAUUGACAGGCAAAAAUUAAAGACGAAAGUCGACUAGCGUAAUAAUCUUCAACAAAGAAAAUAUUAGGUUAAGCAUGAGCUA-ACAUAAAUAUAUAUGAUUAGGU .((((.......(((..........(((....)))(..((((.(((((.((((.....))))...))))).))))..)....))).-.((((......)))).)))).. ( -19.50) >DroSec_CAF1 31808 108 - 1 GCUAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUAGCGUAAUAAUCUUUACCAGAGAAAAUAUUAGGCUAAGCAUGAGCUA-ACAUAAAUAUAUAUGAAUAGGU .(((........(((..........(((....)))(..((((.(((((.((((.....))))...))))).))))..)....))).-.((((......))))..))).. ( -21.80) >DroSim_CAF1 30936 108 - 1 GCAAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUAGCGUAAUAAUCUUUACCAGAGAAAAUAUUAGGCUAAGCAUGAGCUA-ACAUAAAUAUAUAUGAAUAGGU ............(((..........(((....)))(..((((.(((((.((((.....))))...))))).))))..)....))).-.((((......))))....... ( -20.90) >DroEre_CAF1 42208 95 - 1 GCCAAAUUGACAGGCAAAAAUUACAGACGGAAGUCGACCUGC-UGAUAAUCUUUAACAGAGAAAAUAUCAGGCUAGUCACGAGCUAAUCACUAAUA------------- .......(((..(((..........(((....)))(((..((-(((((.((((.....))))...))))).))..)))....)))..)))......------------- ( -22.70) >DroYak_CAF1 33262 99 - 1 GCUAAAUUGACAGGCGGGAAUAGAAGACGGAAGUCGACCUGC-UAAUAAUCUUUAGCUAAGAAAAUAUAAAGCUAAUUGUGAGCUA-UCAUAAAUAUACAU-------- ((((((......((((((.......(((....)))..)))))-).......))))))...................((((((....-))))))........-------- ( -21.72) >consensus GCUAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUAGCGUAAUAAUCUUUAACAGAGAAAAUAUUAGGCUAAGCAUGAGCUA_ACAUAAAUAUAUAUGA_UAGGU ............(((..........(((....)))(..((((.(((((.((((.....))))...))))).))))..)....)))........................ (-15.42 = -15.62 + 0.20)


| Location | 11,773,805 – 11,773,924 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.57 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11773805 119 - 22407834 UUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCUAAAUUGACAGGCAAAAAUUAAAGACGAAAGUCGACUAGCGUAAUAAUCUUCAACAAAGAAAAUAUUAGGUUAAGCAU (((((......(((.....)))..(((((.........)))))........)))))........(((....)))(..((((.(((((.((((.....))))...))))).))))..).. ( -23.50) >DroSec_CAF1 31836 119 - 1 UUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGAGCUAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUAGCGUAAUAAUCUUUACCAGAGAAAAUAUUAGGCUAAGCAU (((((....(((...........))).....((((...........)))).)))))........(((....)))(..((((.(((((.((((.....))))...))))).))))..).. ( -24.80) >DroSim_CAF1 30964 119 - 1 UUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCAAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUAGCGUAAUAAUCUUUACCAGAGAAAAUAUUAGGCUAAGCAU (((((....(((...........)))(((.........)))..........)))))........(((....)))(..((((.(((((.((((.....))))...))))).))))..).. ( -26.00) >DroEre_CAF1 42224 118 - 1 UUGCCACAACAUAUUACUAAUACAUGGCUAAGUCACGGGGCCAAAUUGACAGGCAAAAAUUACAGACGGAAGUCGACCUGC-UGAUAAUCUUUAACAGAGAAAAUAUCAGGCUAGUCAC (((((......(((.....)))..(((((..(...)..)))))........)))))........(((....)))(((..((-(((((.((((.....))))...))))).))..))).. ( -28.20) >DroYak_CAF1 33282 118 - 1 UUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCUAAAUUGACAGGCGGGAAUAGAAGACGGAAGUCGACCUGC-UAAUAAUCUUUAGCUAAGAAAAUAUAAAGCUAAUUGU .....((((..((((.........(((((.........)))))........((((((.......(((....)))..)))))-)))))....((((((............)))))))))) ( -22.90) >consensus UUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCUAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUAGCGUAAUAAUCUUUAACAGAGAAAAUAUUAGGCUAAGCAU (((((......(((.....)))..(((((.........)))))........)))))........(((....)))(..((((.(((((.((((.....))))...))))).))))..).. (-20.66 = -20.70 + 0.04)

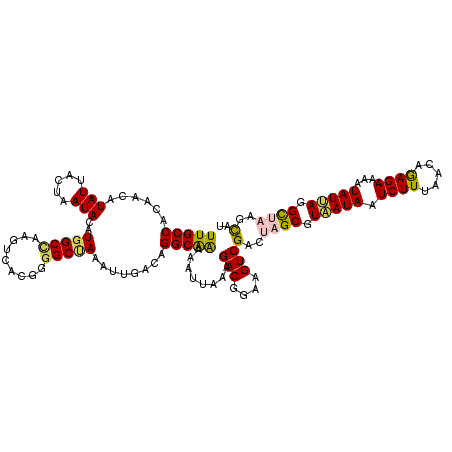
| Location | 11,773,845 – 11,773,962 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 94.46 |
| Mean single sequence MFE | -25.39 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.64 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11773845 117 - 22407834 GCUAAACAAUGGCCAA-AACUUUCGAUGGUUUCGAACUUUUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCUAAAUUGACAGGCAAAAAUUAAAGACGAAAGUCGACUA (((...(((((((.((-((..(((((.....))))).)))))))...................(((((.........))))).))))...)))..........(((....)))..... ( -25.30) >DroSec_CAF1 31876 117 - 1 GCUAAACAAUGGCCAA-AACUUUCGAUGGUUUCGAACUUUUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGAGCUAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUA .........((((((.-....(((((.....)))))....((......)).............))))))((((.....(((.........)))..........(((....))))))). ( -24.90) >DroSim_CAF1 31004 117 - 1 GCUAAACAAUGGCCAA-AACUUUCGAUGGUUUCGAACUUUUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCAAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUA ((((.....))))...-..((.(((((.((..((.((((..((((..................)))).))))..))..))...))))).))............(((....)))..... ( -24.77) >DroEre_CAF1 42263 117 - 1 GCUAAACAAUGGCCAA-AACUUUCGAUUGUUUCGAACUUUUGCCACAACAUAUUACUAAUACAUGGCUAAGUCACGGGGCCAAAUUGACAGGCAAAAAUUACAGACGGAAGUCGACCU (((...((((((((..-.....((((.....))))((((..((((..................)))).)))).....)))))..)))...)))..........(((....)))..... ( -27.07) >DroYak_CAF1 33321 118 - 1 GCUAAACAAUGGCCAAAAACUUUCGAUUGUUUCGAACUUUUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCUAAAUUGACAGGCGGGAAUAGAAGACGGAAGUCGACCU (((...(((((((.((((...(((((.....))))).)))))))...................(((((.........))))).))))...)))..........(((....)))..... ( -24.90) >consensus GCUAAACAAUGGCCAA_AACUUUCGAUGGUUUCGAACUUUUGCCACAACAUAUUACUAAUACAUGGCCAAGUCACGGGGCUAAAUUGACAGGCAAAAAUUAAAGACGGAAGUCGACUA (((......((((((......(((((.....)))))....((......)).............)))))).((((...........)))).)))..........(((....)))..... (-23.80 = -23.64 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:55 2006