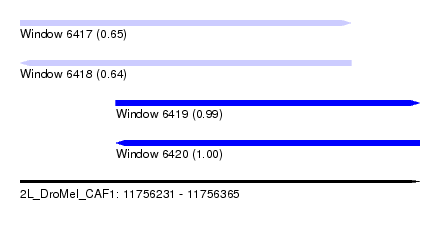
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,756,231 – 11,756,365 |
| Length | 134 |
| Max. P | 0.998018 |

| Location | 11,756,231 – 11,756,342 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -22.35 |
| Energy contribution | -24.38 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11756231 111 + 22407834 G-----GUUGGACGAAGCCAAAAAGGCUGCAGG---CAAACGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGAGAG-GAGGAUGGUGUUGGUGUGCAAU (-----(((......)))).....(((((((.(---(....))..)).))))).((((((((((((....(((((((....)))))))...(....)-.....))))))))))))..... ( -32.20) >DroSec_CAF1 14402 112 + 1 G-----GUUGAACGAAGCCAAAAAGGCUGCAGG---CAAACGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGUAGGGUGGGUGGUGUUGGUGUGCAAU (-----(((......)))).....(((((((.(---(....))..)).))))).((((((((((((...((((((((....))))))).(((......)))).))))))))))))..... ( -34.00) >DroSim_CAF1 13296 111 + 1 G-----GUUGAACGAAGCCAAAAAGGCUGCAGG---CAAACGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGUAG-GUGGGUGGUGUUGGUGUGCAAU (-----(((......)))).....(((((((.(---(....))..)).))))).((((((((((((...((((((((....))))))).(((.....-)))).))))))))))))..... ( -33.80) >DroEre_CAF1 21831 116 + 1 GGUGGUGGUGGGCAAAGCCAAAAAGGCUGCAGG---CAAAUGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGAG-GAGGGUGGUGUUGGUGUGCAAU (((..((.....))..)))......((((((..---....))))))....((..((((((((((((...((((((((....)))))))...(....)-..)..))))))))))))))... ( -34.10) >DroYak_CAF1 15347 115 + 1 GGAGAUGGUGGGAAAAGCCAAAAAGGCUGCAGG---CAAAUGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGAG-G-GUGUGGUGUUGGUGUGCAAU ...((((.(((......))).....((((((..---....)))))).........)))).(((((((((((((((((....)))))))((((.....-.-)))).))))))))))..... ( -34.40) >DroAna_CAF1 16817 96 + 1 -----------GCAUUGCCAAAAAGGCUGCAGGCUGCAAAUGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGGC-AGGGGCGGCA------------ -----------(((..(((.....))))))..(((((...(((.(((...((((....))))........(((((((....))))))).)))...))-)...))))).------------ ( -29.40) >consensus G_____GUUGGACAAAGCCAAAAAGGCUGCAGG___CAAACGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGAG_GAGGGUGGUGUUGGUGUGCAAU ...............((((.....))))((............(((((........)))))(((((((((((((((((....))))))).(((..........)))))))))))))))... (-22.35 = -24.38 + 2.03)

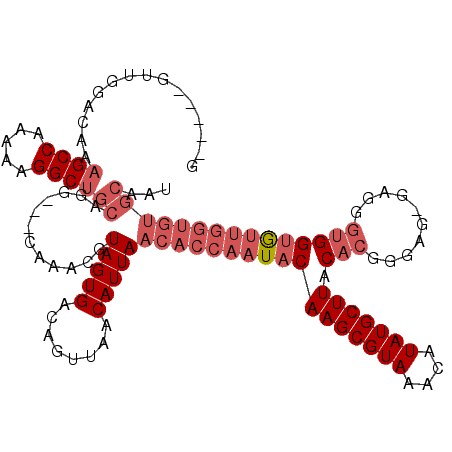

| Location | 11,756,231 – 11,756,342 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.28 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11756231 111 - 22407834 AUUGCACACCAACACCAUCCUC-CUCUCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACGUUUG---CCUGCAGCCUUUUUGGCUUCGUCCAAC-----C .....(((..(((((((.....-..(..((((((((....))))))))..)...)))))))..)))...............(((---..((.((((.....)))).))..))).-----. ( -24.90) >DroSec_CAF1 14402 112 - 1 AUUGCACACCAACACCACCCACCCUACCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACGUUUG---CCUGCAGCCUUUUUGGCUUCGUUCAAC-----C .....(((..(((((((.......(((.((((((((....))))))))..))).)))))))..)))...............(((---..((.((((.....)))).))..))).-----. ( -23.80) >DroSim_CAF1 13296 111 - 1 AUUGCACACCAACACCACCCAC-CUACCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACGUUUG---CCUGCAGCCUUUUUGGCUUCGUUCAAC-----C .....(((..(((((((.....-.(((.((((((((....))))))))..))).)))))))..)))...............(((---..((.((((.....)))).))..))).-----. ( -24.40) >DroEre_CAF1 21831 116 - 1 AUUGCACACCAACACCACCCUC-CUCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACAUUUG---CCUGCAGCCUUUUUGGCUUUGCCCACCACCACC ..((((....(((((((.....-.....((((((((....))))))))......)))))))(((((...........)))))..---..))))(((.....)))................ ( -23.59) >DroYak_CAF1 15347 115 - 1 AUUGCACACCAACACCACAC-C-CUCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACAUUUG---CCUGCAGCCUUUUUGGCUUUUCCCACCAUCUCC ..((((....(((((((...-.-.....((((((((....))))))))......)))))))(((((...........)))))..---..))))(((.....)))................ ( -24.16) >DroAna_CAF1 16817 96 - 1 ------------UGCCGCCCCU-GCCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACAUUUGCAGCCUGCAGCCUUUUUGGCAAUGC----------- ------------(((((...((-((...((((((((....)))))))).......(((((.(((((...........))))).))).)).))))......)))))....----------- ( -25.10) >consensus AUUGCACACCAACACCACCCAC_CUCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACAUUUG___CCUGCAGCCUUUUUGGCUUCGCCCAAC_____C ..((((....(((((((...........((((((((....))))))))......)))))))(((((...........))))).......))))(((.....)))................ (-20.01 = -20.35 + 0.33)


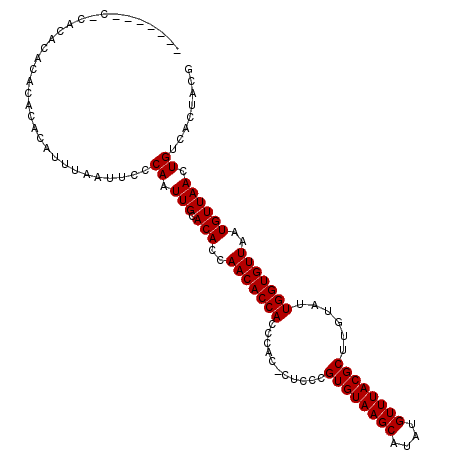
| Location | 11,756,263 – 11,756,365 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.34 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -24.70 |
| Energy contribution | -24.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11756263 102 + 22407834 CGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGAGAG-GAGGAUGGUGUUGGUGUGCAAUUGGGAAUUAAAUGUGUGUGUGUG----------- ..((((..(((((.((((((((((((....(((((((....)))))))...(....)-.....))))))))))))..)))))...))))..............----------- ( -28.00) >DroSec_CAF1 14434 105 + 1 CGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGUAGGGUGGGUGGUGUUGGUGUGCAAUUGGGAAUUAGAUGUGUG--UGUGUGAG------- ..((((..(((((.((((((((((((...((((((((....))))))).(((......)))).))))))))))))..)))))...))))........--........------- ( -29.80) >DroSim_CAF1 13328 106 + 1 CGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGUAG-GUGGGUGGUGUUGGUGUGCAAUUGGGAAUUAAAUGUGUGUGUGUGUGAG------- ..((((..(((((.((((((((((((...((((((((....))))))).(((.....-)))).))))))))))))..)))))...))))..................------- ( -29.60) >DroEre_CAF1 21868 101 + 1 UGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGAG-GAGGGUGGUGUUGGUGUGCAAUUGGGAAUUUAACGUGUGUGUGU------------ ........(((((.((((((((((((...((((((((....)))))))...(....)-..)..))))))))))))..)))))....................------------ ( -26.80) >DroYak_CAF1 15384 112 + 1 UGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGAG-G-GUGUGGUGUUGGUGUGCAAUUGGGAAUUUAAUGUGUGUGUGUGUGUGUCACACA .((.((((((....((((..(((((((((((((((((....)))))))((((.....-.-)))).))))))))))((((((((....))))).)))..))))...)))))))). ( -34.90) >consensus CGUAGUGACAGUUAACAUUAACACCAAUACAAGCGUAAACAUAUGCUUACACGGGAG_GAGGGUGGUGUUGGUGUGCAAUUGGGAAUUAAAUGUGUGUGUGUGUG_G_______ ........(((((.((((((((((((...((((((((....)))))))...(....)...)..))))))))))))..)))))................................ (-24.70 = -24.70 + -0.00)



| Location | 11,756,263 – 11,756,365 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.34 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -16.77 |
| Energy contribution | -16.77 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11756263 102 - 22407834 -----------CACACACACACAUUUAAUUCCCAAUUGCACACCAACACCAUCCUC-CUCUCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACG -----------.....................((.(((.(((..(((((((.....-..(..((((((((....))))))))..)...)))))))..)))))).))........ ( -18.60) >DroSec_CAF1 14434 105 - 1 -------CUCACACA--CACACAUCUAAUUCCCAAUUGCACACCAACACCACCCACCCUACCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACG -------........--...............((.(((.(((..(((((((.......(((.((((((((....))))))))..))).)))))))..)))))).))........ ( -17.50) >DroSim_CAF1 13328 106 - 1 -------CUCACACACACACACAUUUAAUUCCCAAUUGCACACCAACACCACCCAC-CUACCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACG -------.............(((.(((((.....((((.(((((((..........-.(((.((((((((....))))))))..))))))))))))))))))).)))....... ( -18.20) >DroEre_CAF1 21868 101 - 1 ------------ACACACACACGUUAAAUUCCCAAUUGCACACCAACACCACCCUC-CUCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACA ------------......(((.(((((.......((((.(((((((.((.......-.....((((((((....))))))))..)).))))))))))).))))))))....... ( -18.84) >DroYak_CAF1 15384 112 - 1 UGUGUGACACACACACACACACAUUAAAUUCCCAAUUGCACACCAACACCACAC-C-CUCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACA ((((((.....))))))...............((.(((.(((..(((((((...-.-.....((((((((....))))))))......)))))))..)))))).))........ ( -23.96) >consensus _______C_CACACACACACACAUUUAAUUCCCAAUUGCACACCAACACCACCCAC_CUCCCGUGUAAGCAUAUGUUUACGCUUGUAUUGGUGUUAAUGUUAACUGUCACUACG ................................((.(((.(((..(((((((...........((((((((....))))))))......)))))))..)))))).))........ (-16.77 = -16.77 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:41 2006