| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,751,455 – 11,751,563 |
| Length | 108 |
| Max. P | 0.773695 |

| Location | 11,751,455 – 11,751,563 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -13.48 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11751455 108 - 22407834 UUUCCAUCUCCCCAUCCGCAUCCGCAUCCCCAUUCACUUCCUUUUUUCUUUUGC---GAUUCGAGGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAAACGACA ......((((.((....((....))............((((((...((......---))...))))))......((((((........))))))...)).))))....... ( -17.10) >DroSec_CAF1 9722 85 - 1 -----------------------CCAUCUCCAUCCACUUCCUCUUUCUUUUUGC---GAUUCGAGGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAAACGACA -----------------------...((((...((..((((((..((.......---))...))))))......((((((........))))))...)).))))....... ( -15.00) >DroSim_CAF1 8645 85 - 1 -----------------------CCAUCUCCAUCCACUUCCUUUUUCUUUUUGC---GAUUCGAGGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAAACGACA -----------------------...((((...((..((((((..((.......---))...))))))......((((((........))))))...)).))))....... ( -13.20) >DroEre_CAF1 16878 102 - 1 UUU------CCCCAUCUCCAUCCCCAUCCCCAUCCACUUCCUUUUUCUUUUUGC---GAUUCGAGGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAAACGACA (((------(.((........................((((((..((.......---))...))))))......((((((........))))))...)).))))....... ( -12.50) >DroYak_CAF1 10154 102 - 1 UUU------CCCCAUCUCCAUUCCCAUCUCCAUUCACUUCCAUUUUCUUUUUGC---GAUUCGAGGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAAACGACA ...------.................................(((((((((((.---((((.((((((((((...))))))))))...))))))))))).))))....... ( -11.00) >DroAna_CAF1 11373 91 - 1 ---------------CUUUUUUCCCAUUUCUGG--GU---UUUUAUUUUUUUGCACCAUUUUCAAGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAGACGACA ---------------.......((((....)))--).---......((((((((.........((....))...((((((........))))))....))))))))..... ( -12.10) >consensus _______________CU_CAU_CCCAUCUCCAUCCACUUCCUUUUUCUUUUUGC___GAUUCGAGGAAAUUUUAUGAAUUUUCUUAUCAAUUCAAAAGGCGAGAAACGACA .........................................((((.(((((((....(((..((((((((((...)))))))))))))....))))))).))))....... (-10.00 = -10.50 + 0.50)


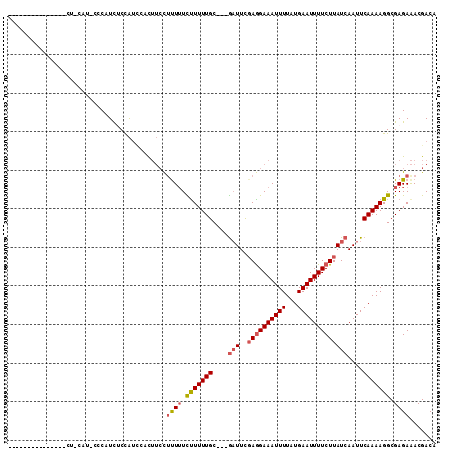
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:35 2006