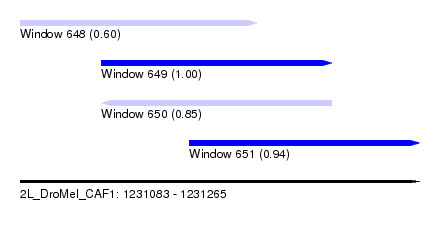
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,231,083 – 1,231,265 |
| Length | 182 |
| Max. P | 0.996801 |

| Location | 1,231,083 – 1,231,191 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.38 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -22.49 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1231083 108 + 22407834 AUACGAAGUACACAA--GCACAAACAACGCCGGAAC-GGAAACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACUUUC---------CCGGU ...............--...........(((((...-(((((((((....)).......(((((....)))))....))))))).(((((((......))))))).---------))))) ( -32.00) >DroSec_CAF1 8433 109 + 1 AUACGAAGUACAACAACACACAAACAACGCCGGAGC-GGAAACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAUCCAACCACUUUC---------CCGG- .............................((((.(.-(((((((((....)).......(((((....)))))....))))))))(((((((......))))))).---------))))- ( -30.20) >DroSim_CAF1 4229 104 + 1 AUACGAAGUACACAA--GCACAAACAACGCCGGAGCCGGAAACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACUUUC-------------- .(((...........--............(((....)))...((((....)).)).)))(((((....)))))..(((....)))(((((((......))))))).-------------- ( -24.00) >DroYak_CAF1 4305 117 + 1 AUACGAAGUACACAA--GCACAAACAACGCCACUGC-GGAAACCGCAGCAGCAGGAGUAGACGUAGCAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACCCUUCCACUCUGCCCGUU ...............--........(((((..((((-((...))))))..)).((.(((((.((...........(((....)))..(((((......))))).....)))))))))))) ( -33.10) >consensus AUACGAAGUACACAA__GCACAAACAACGCCGGAGC_GGAAACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACUUUC_________CCGG_ .....................................(((((((((....)).......(((((....)))))....))))))).(((((((......)))))))............... (-22.49 = -22.80 + 0.31)

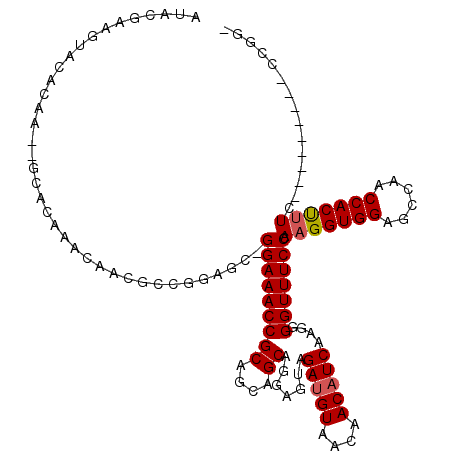
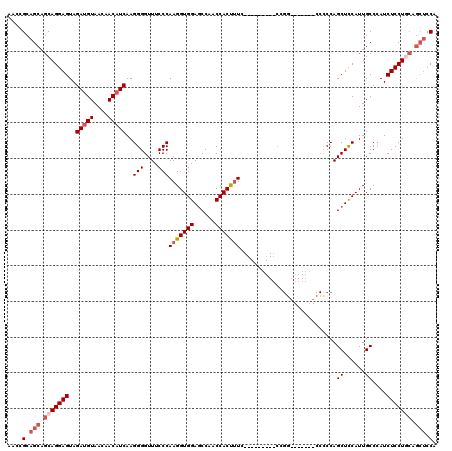
| Location | 1,231,120 – 1,231,225 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -25.54 |
| Energy contribution | -27.35 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1231120 105 + 22407834 AACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACUUUC---------CCGGU------CCCCCAGCUCCAUUGCCCAUCUCCUCCAGCUCCA .......(.(((.((((.(((((............((((..((..(((((((......))))))).---------..)).------.)))).((......)).))))).)))).))).). ( -34.30) >DroSec_CAF1 8472 104 + 1 AACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAUCCAACCACUUUC---------CCGG-------CCCCCAGCUCCAUUGCCAAUCUCCUGCAGCUCCA ....(.(((.(((((((..(((((....)))))..((((((....(((((((......))))))).---------..))-------))))..((......))....))))))).))).). ( -38.70) >DroSim_CAF1 4267 98 + 1 AACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACUUUC----------------------CCCAGCUCCAUUGCCCAUCUCCUGCAGCUCCA ....(.(((.(((((((..(((((....)))))..((((((....(((((((......))))))).----------------------...)))))).........))))))).))).). ( -36.50) >DroYak_CAF1 4342 114 + 1 AACCGCAGCAGCAGGAGUAGACGUAGCAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACCCUUCCACUCUGCCCGUUCCCAAGUCACCAGCUUCAUUGCCCAUCUCCU------CCA ....((....)).((((.(((....((((......((((.....((..((((((..........))))))..)).....))))((((.....))))..))))...))).))------)). ( -27.80) >consensus AACCGCAGCAGCAGGAGUAGAUGUAACAACAUCAAGGGGUUUCCCAAGGUGGAGCCAACCACUUUC_________CCGG_______CCCCCAGCUCCAUUGCCCAUCUCCUGCAGCUCCA ....(.(((.(((((((..(((((....)))))..(((....)))(((((((......)))))))...........................((......))....))))))).))).). (-25.54 = -27.35 + 1.81)

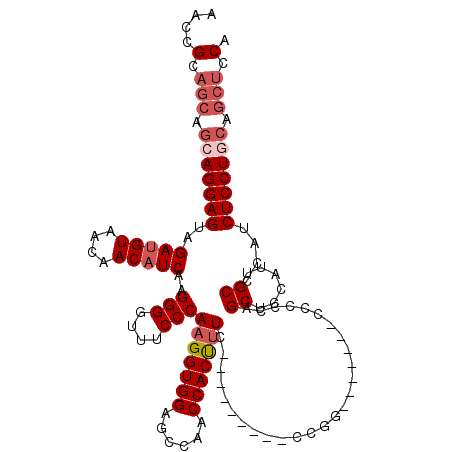
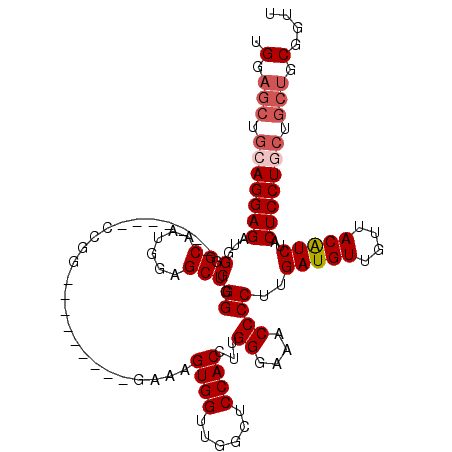
| Location | 1,231,120 – 1,231,225 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -29.64 |
| Energy contribution | -30.95 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1231120 105 - 22407834 UGGAGCUGGAGGAGAUGGGCAAUGGAGCUGGGGG------ACCGG---------GAAAGUGGUUGGCUCCACCUUGGGAAACCCCUUGAUGUUGUUACAUCUACUCCUGCUGCUGCGGUU .(.(((.((((.((((((((((((...(.(((((------.((((---------(...((((......)))))))))....))))).).))))))).))))).)))).))).)....... ( -41.80) >DroSec_CAF1 8472 104 - 1 UGGAGCUGCAGGAGAUUGGCAAUGGAGCUGGGGG-------CCGG---------GAAAGUGGUUGGAUCCACCUUGGGAAACCCCUUGAUGUUGUUACAUCUACUCCUGCUGCUGCGGUU .(.(((.(((((((...(((......)))((((.-------((((---------(...((((......)))))))))....))))..(((((....)))))..))))))).))).).... ( -42.70) >DroSim_CAF1 4267 98 - 1 UGGAGCUGCAGGAGAUGGGCAAUGGAGCUGGG----------------------GAAAGUGGUUGGCUCCACCUUGGGAAACCCCUUGAUGUUGUUACAUCUACUCCUGCUGCUGCGGUU .(.(((.(((((((..((....((((((..(.----------------------........)..))))))....((....))))..(((((....)))))..))))))).))).).... ( -40.80) >DroYak_CAF1 4342 114 - 1 UGG------AGGAGAUGGGCAAUGAAGCUGGUGACUUGGGAACGGGCAGAGUGGAAGGGUGGUUGGCUCCACCUUGGGAAACCCCUUGAUGUUGCUACGUCUACUCCUGCUGCUGCGGUU .((------((.(((((((((((...(((.((..(...)..)).))).....((.(((((((......)))))))((....)))).....)))))).))))).))))(((....)))... ( -41.40) >consensus UGGAGCUGCAGGAGAUGGGCAAUGGAGCUGGGGG_______CCGG_________GAAAGUGGUUGGCUCCACCUUGGGAAACCCCUUGAUGUUGUUACAUCUACUCCUGCUGCUGCGGUU .(.(((.(((((((...(((......)))((...........................((((......))))...((....))))..(((((....)))))..))))))).))).).... (-29.64 = -30.95 + 1.31)

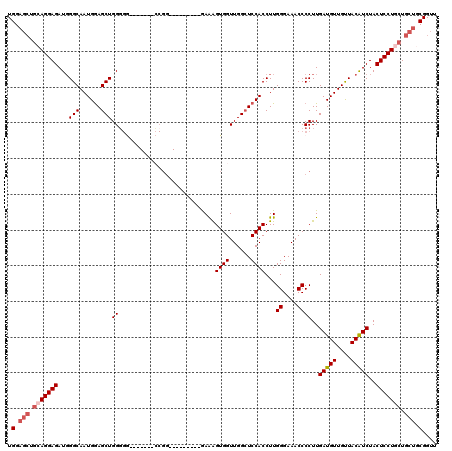
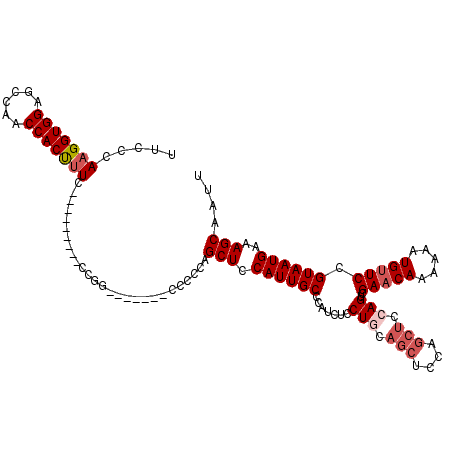
| Location | 1,231,160 – 1,231,265 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.49 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -18.41 |
| Energy contribution | -19.98 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1231160 105 + 22407834 UUCCCAAGGUGGAGCCAACCACUUUC---------CCGGU------CCCCCAGCUCCAUUGCCCAUCUCCUCCAGCUCCAGCUCCAGUCGAACAAAAAAUGUUCCGUAAUGAAAGCAAUU .......((.((..((..........---------..)).------.)))).(((.((((((.......((..(((....)))..))..(((((.....))))).))))))..))).... ( -21.70) >DroSec_CAF1 8512 104 + 1 UUCCCAAGGUGGAUCCAACCACUUUC---------CCGG-------CCCCCAGCUCCAUUGCCAAUCUCCUGCAGCUCCAGCUCCAGUCGAACAAAAAAUGUUCCGUAAUGAAAGCAAUU ..((.(((((((......))))))).---------..))-------......(((.((((((.......(((.(((....))).)))..(((((.....))))).))))))..))).... ( -24.70) >DroSim_CAF1 4307 98 + 1 UUCCCAAGGUGGAGCCAACCACUUUC----------------------CCCAGCUCCAUUGCCCAUCUCCUGCAGCUCCAGCUCCAGUCGAACAAAAAAUGUUCCGUAAUGAAAGCAAUU .....(((((((......))))))).----------------------....(((.((((((.......(((.(((....))).)))..(((((.....))))).))))))..))).... ( -24.20) >DroYak_CAF1 4382 114 + 1 UUCCCAAGGUGGAGCCAACCACCCUUCCACUCUGCCCGUUCCCAAGUCACCAGCUUCAUUGCCCAUCUCCU------CCAGCUCCAGUCGAACAAAAAAUGCUCAGUAAUGAAAGCAAUU ....((..((((((..........))))))..))..................((((((((((.........------...((....)).((.((.....)).)).)))))).)))).... ( -17.40) >consensus UUCCCAAGGUGGAGCCAACCACUUUC_________CCGG_______CCCCCAGCUCCAUUGCCCAUCUCCUGCAGCUCCAGCUCCAGUCGAACAAAAAAUGUUCCGUAAUGAAAGCAAUU .....(((((((......)))))))...........................(((.((((((.......(((.(((....))).)))..(((((.....))))).))))))..))).... (-18.41 = -19.98 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:17 2006