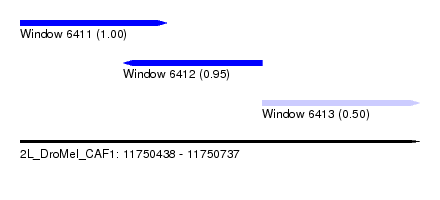
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,750,438 – 11,750,737 |
| Length | 299 |
| Max. P | 0.999971 |

| Location | 11,750,438 – 11,750,548 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 93.44 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.95 |
| SVM decision value | 5.05 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11750438 110 + 22407834 UGCCACCUCGAGUCAUCCUUUUAAUUAUGUAACACGCGCCACCACAGGAGUGUUGCCUUACGCACUUCGGUCUUCGGUCUGCGGUCCUUUUUGGUGGCAAACAUUU-------UCCA (((((((..(((................(((((((...((......)).)))))))....((((..(((.....)))..))))...)))...))))))).......-------.... ( -32.50) >DroSec_CAF1 8760 110 + 1 UGCCACCUCGAGCCAUCCUUUUAAUUAUGUAACACGCGCCACCACAGGAGUGUUGCCUUACGCACUUCGGUCUUCGGUCUGCGGUCCUUUUUGGUGGCAAAAAUUU-------UCCA (((((((..(..((..............(((((((...((......)).))))))).....(((..(((.....)))..)))))..).....))))))).......-------.... ( -32.60) >DroSim_CAF1 7630 110 + 1 UGCCACCUCGAGCCAUCCUUUUAAUUAUGUAACACGCGCCACCACAGGAGUGUUGCCUUACGCACUUCGGUCUUCGGUCUGCGGUCCUUUUUGGUGGCAAACAUUU-------UCCA (((((((..(..((..............(((((((...((......)).))))))).....(((..(((.....)))..)))))..).....))))))).......-------.... ( -32.60) >DroEre_CAF1 15897 117 + 1 UGCCACCUCAAGCCAUCUUUUUAAUUAUGUAACACGCGCCACCACUGGAGUGUUGCCUUACGCACUUCGGUCUUCGGUCUGCGGCUCUUUUUGGUGGCAAACAUUUUCCAUUUUCCA (((((((...((((..............(((((((...(((....))).))))))).....(((..(((.....)))..)))))))......))))))).................. ( -35.80) >DroYak_CAF1 9157 109 + 1 UGCCACCUCGAGCCAUCUUUUUAAUUAUGUAACACGCGCCGCCACUGGAGUGUUGCCUUACGCACUUCGGUCUUCGGUCUGCGGUUCUUUUUGGUGGCAAACAUA--------UCCA (((((((..(((((..............(.....)((((((..((((((((((........))))))))))...))))..))))))).....)))))))......--------.... ( -37.10) >consensus UGCCACCUCGAGCCAUCCUUUUAAUUAUGUAACACGCGCCACCACAGGAGUGUUGCCUUACGCACUUCGGUCUUCGGUCUGCGGUCCUUUUUGGUGGCAAACAUUU_______UCCA (((((((..(((................(((((((...((......)).)))))))....((((..(((.....)))..))))...)))...))))))).................. (-32.32 = -32.16 + -0.16)



| Location | 11,750,515 – 11,750,619 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.56 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -24.45 |
| Energy contribution | -24.27 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11750515 104 - 22407834 AAGAACACGCAACUACUGGCCAACGGAAGUGUGGAAUAUGCCCAGUGGGGGGAAAAAUGUUGGCAGGAAAAUGGAAAAUGUUUGCCACCAAAAAGGACCGCAGA ........((..((((((((((((....)).))).......)))))))((.......((.(((((((...((.....)).))))))).)).......))))... ( -27.65) >DroSec_CAF1 8837 102 - 1 AAGAACACGCAACUACUGGCCAACGGAAGUGUGGAAUAUGCCCAGUGGGGGG--AAAUGUUGGCAGGAAAAUGGAAAAUUUUUGCCACCAAAAAGGACCGCAGA ........((..((((((((((((....)).))).......))))))).((.--......(((((((((.........)))))))))((.....)).))))... ( -28.41) >DroSim_CAF1 7707 102 - 1 AAGAACACGCAACUACUGGCCAACGGAAGUGUGGAAUAUGCCCAGUGGGGGA--AAAUGUUGGCAGGAAAAUGGAAAAUGUUUGCCACCAAAAAGGACCGCAGA ........((..((((((((((((....)).))).......)))))))((..--......(((((((...((.....)).)))))))((.....)).))))... ( -27.31) >DroYak_CAF1 9234 100 - 1 AAGGACACGCAACUACUGGCCAACGGAAGUGCUGAAAAUGCUCAGUGGGG-A--AAGUGGUGGCAGGAAAAUGGA-UAUGUUUGCCACCAAAAAGAACCGCAGA ........((..(((((((...((....))((.......)))))))))((-.--...((((((((((...((...-.)).)))))))))).......))))... ( -28.80) >consensus AAGAACACGCAACUACUGGCCAACGGAAGUGUGGAAUAUGCCCAGUGGGGGA__AAAUGUUGGCAGGAAAAUGGAAAAUGUUUGCCACCAAAAAGGACCGCAGA ........((..(((((((.((((....))........)).)))))))((.......((.(((((((...((.....)).))))))).)).......))))... (-24.45 = -24.27 + -0.19)


| Location | 11,750,619 – 11,750,737 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -19.06 |
| Energy contribution | -17.75 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11750619 118 + 22407834 CAAGUGUGAACUUAAUUCAGAUAAAGCGAAAAUUCUGAUAAGUUUAAUUGCUAUCCUAAAUGGUGACAAAGCGUUUGCAUUUAUAUUCAUGUAUACACAUACACAUGAAUAGCCGUAG .(((((..(((.....(((((............)))))...((((..(..((((.....))))..)..)))))))..))))).(((((((((.((....)).)))))))))....... ( -27.10) >DroSim_CAF1 7809 118 + 1 CACGUGUGAACUUAAUUCAGAUAAAGCGAAAAUUCUGAUAAGUUUAAUUGCUAGCCUAAAUGGUGACAAAGCGUUAGCGCUUAUUUUUAUGUAUACACAUACACAUGAAUAGCCGUAG .(((..((((((((..(((((............)))))))))))))...((((((((...((....)).)).))))))(((.((((...(((((....)))))...)))))))))).. ( -27.90) >DroEre_CAF1 16082 110 + 1 CACGUGUGAACUUAAUUCAGAUAAAGCGAAAACUCUGAUAAGUUUAAUUGUUGGCCUAAAUGGUGACAAAGCGUUUGCACUUAUUUUCAGGGAUA--------UUUGAAUAGCCAUAG .(((..((((((((..(((((............)))))))))))))..)))((((......((((.(((.....)))))))....((((((....--------))))))..))))... ( -20.90) >DroYak_CAF1 9334 110 + 1 CACGUGUGAACUUUAUUCAGAUAAAGCGAAAAUUCUGAUAAGUUUAAUUGCUAUCCUAAAUGGUGACAAAGCGUUUGCACUUGCUUUCAUGGGAU--------UAUGAAUAGCCACAG ...(((..(((.....(((((............)))))...((((..(..((((.....))))..)..)))))))..)))..(((((((((....--------)))))).)))..... ( -24.80) >consensus CACGUGUGAACUUAAUUCAGAUAAAGCGAAAAUUCUGAUAAGUUUAAUUGCUAGCCUAAAUGGUGACAAAGCGUUUGCACUUAUUUUCAUGGAUA________CAUGAAUAGCCAUAG ...((((((((.....(((((............)))))...((((..(..(((.......)))..)..)))))))))))).....((((((............))))))......... (-19.06 = -17.75 + -1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:34 2006