
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,750,117 – 11,750,240 |
| Length | 123 |
| Max. P | 0.958097 |

| Location | 11,750,117 – 11,750,217 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.31 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -31.22 |
| Energy contribution | -31.66 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.754982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
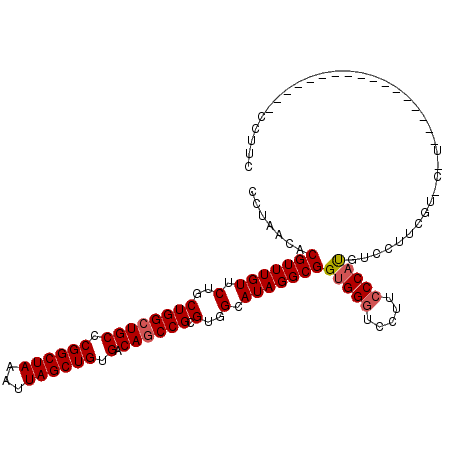
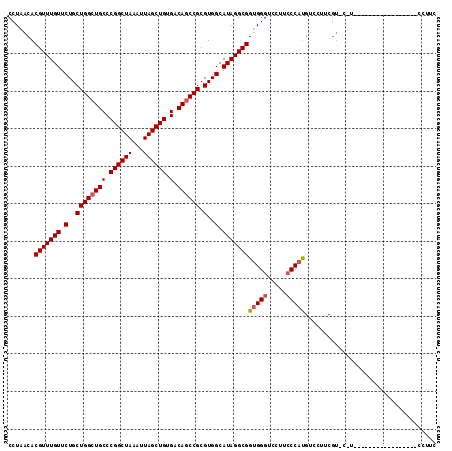
>2L_DroMel_CAF1 11750117 100 - 22407834 CCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUUCCCAUGUCCUUCGUUCUUCGU--------------CCUUC ...(((.(((((((.(..((((((((.((((((...)))))).).)))))).)..).)))))))(((((.....))))).......)))......--------------..... ( -34.20) >DroSec_CAF1 8460 79 - 1 CCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUUCUGCCCAU----------------------------------- .......(((((((.(..((((((((.((((((...)))))).).)))))).)..).)))))))(((((.....)))))----------------------------------- ( -33.90) >DroSim_CAF1 7317 92 - 1 CCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUGCCCAUGUCCUUCGUCCAU---------------------- .......(((((((.(..((((((((.((((((...)))))).).)))))).)..).)))))))(((((.....))))).............---------------------- ( -34.60) >DroEre_CAF1 15595 93 - 1 CCUAACACGUUUGUUCUGCUGGAUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGCGGGUCCUUGCCAUGUCCUUCGU---------------------CCUUC ....(((.(((.(((..(((((....)))))..))).))))))(((((..((((((((..(((.....)))..))))))))..)).))---------------------).... ( -29.40) >DroYak_CAF1 8833 114 - 1 CCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUUCCCACGUCCUUCGUCCUUCGUCCUUCGUCCUUCGUCCUUC .......(((((((.(..((((((((.((((((...)))))).).)))))).)..).)))))))(((((.....)))))......((.....((.....)).....))...... ( -36.10) >consensus CCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUUCCCAUGUCCUUCGU_C_U_________________CCUUC .......(((((((.(..((((((((.((((((...)))))).).)))))).)..).)))))))(((((.....)))))................................... (-31.22 = -31.66 + 0.44)

| Location | 11,750,138 – 11,750,240 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.98 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
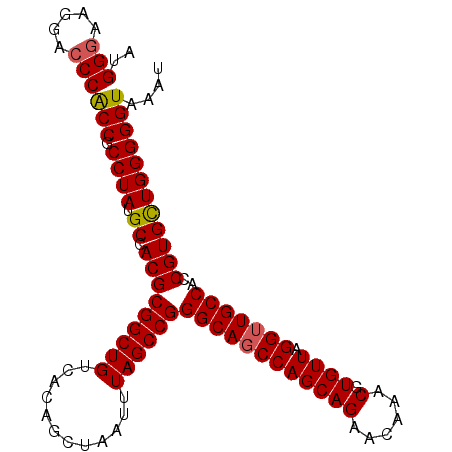
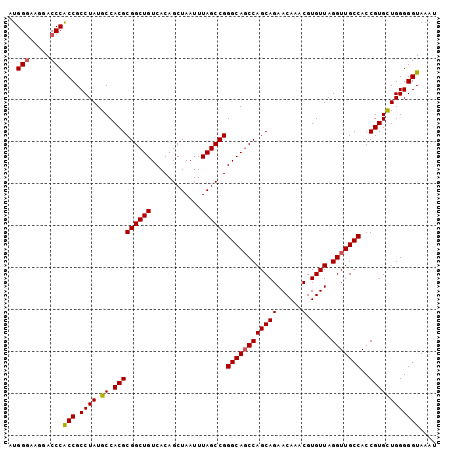
>2L_DroMel_CAF1 11750138 102 + 22407834 AUGGGAAGGACCCACCGCCUAUGCCACGCGGCUGUCACAGCUAAUUUAGCCGGGCAGCCAGCAGAACAAACGUGUUAGGUUGCCACCGUGCUGGGGGUAAAU ..(((.....)))(((.((((.((.(((((((((............))))))((((((((((((......).)))).)))))))..)))))))))))).... ( -38.90) >DroSec_CAF1 8460 102 + 1 AUGGGCAGAACCCACCGCCUAUGCCACGCGGCUGUCACAGCUAAUUUAGCCGGGCAGCCAGCAGAACAAACGUGUUAGGUUGCCACCGUGCUGGGGGUAAAU ..(((.....)))(((.((((.((.(((((((((............))))))((((((((((((......).)))).)))))))..)))))))))))).... ( -38.90) >DroSim_CAF1 7330 102 + 1 AUGGGCAGGACCCACCGCCUAUGCCACGCGGCUGUCACAGCUAAUUUAGCCGGGCAGCCAGCAGAACAAACGUGUUAGGUUGCCACCGUGCUGGGGGUAAAU ..(((.....)))(((.((((.((.(((((((((............))))))((((((((((((......).)))).)))))))..)))))))))))).... ( -38.90) >DroEre_CAF1 15609 102 + 1 AUGGCAAGGACCCGCCGCCUAUGCCACGCGGCUGUCACAGCUAAUUUAGCCGGGCAUCCAGCAGAACAAACGUGUUAGGUUGCCACCGUGCUGGGGGUAAAU ..(((........)))(((((((((...((((((............)))))))))))((((((.((((....)))).(((....))).)))))))))).... ( -34.50) >DroYak_CAF1 8868 102 + 1 GUGGGAAGGACCCACCGCCUAUGCCACGCGGCUGUCACAGCUAAUUUAGCCGGGCAGCCAGCAGAACAAACGUGUUAGGUUGCCACCGUGUUGGGGGUAAAU (((((.....))))).((((....((((((((((............))))))((((((((((((......).)))).)))))))..))))....)))).... ( -39.00) >consensus AUGGGAAGGACCCACCGCCUAUGCCACGCGGCUGUCACAGCUAAUUUAGCCGGGCAGCCAGCAGAACAAACGUGUUAGGUUGCCACCGUGCUGGGGGUAAAU ..(((.....)))(((.((((.((.(((((((((............))))))((((((((((((......).)))).)))))))..)))))))))))).... (-36.90 = -36.98 + 0.08)

| Location | 11,750,138 – 11,750,240 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -35.40 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11750138 102 - 22407834 AUUUACCCCCAGCACGGUGGCAACCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUUCCCAU ...............((..(..(((((...(((((((.(..((((((((.((((((...)))))).).)))))).)..).))))))).)))))..)..)).. ( -37.10) >DroSec_CAF1 8460 102 - 1 AUUUACCCCCAGCACGGUGGCAACCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUUCUGCCCAU ...............((..(.((((((...(((((((.(..((((((((.((((((...)))))).).)))))).)..).))))))).)))))))..))... ( -41.20) >DroSim_CAF1 7330 102 - 1 AUUUACCCCCAGCACGGUGGCAACCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUGCCCAU ....(((........)))(((((((((...(((((((.(..((((((((.((((((...)))))).).)))))).)..).))))))).)))))..))))... ( -39.90) >DroEre_CAF1 15609 102 - 1 AUUUACCCCCAGCACGGUGGCAACCUAACACGUUUGUUCUGCUGGAUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGCGGGUCCUUGCCAU ................((((((((((..((((((.(((..(((((....)))))..))).))).)))...(((((.((...)).)))))))))...)))))) ( -35.30) >DroYak_CAF1 8868 102 - 1 AUUUACCCCCAACACGGUGGCAACCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUUCCCAC ...............((..(..(((((...(((((((.(..((((((((.((((((...)))))).).)))))).)..).))))))).)))))..)..)).. ( -37.10) >consensus AUUUACCCCCAGCACGGUGGCAACCUAACACGUUUGUUCUGCUGGCUGCCCGGCUAAAUUAGCUGUGACAGCCGCGUGGCAUAGGCGGUGGGUCCUUCCCAU ....((((((.....(((....)))......((((((.(..((((((((.((((((...)))))).).)))))).)..).)))))))).))))......... (-35.40 = -35.60 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:31 2006