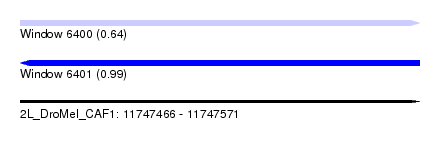
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,747,466 – 11,747,571 |
| Length | 105 |
| Max. P | 0.985146 |

| Location | 11,747,466 – 11,747,571 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.78 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11747466 105 + 22407834 UCAGUUCAGCGAGAACUCUGAGGCAGUCAGCUAUGAACAUGUCCUUGAGACAUGUAAGAGAACUGAGAGCGGCACUCGAAAAAAAA-AGUUGGGAAAAACCAUUUG------ ((((((((((..((.(((....).)))).)))....(((((((.....)))))))....)))))))(((.....))).........-...(((......)))....------ ( -25.00) >DroSec_CAF1 5919 104 + 1 UCA-----GUGAGAACUCCGAGGCAGUCAGCUAUGAACAUGUCCUUGAGACAUGUAAGAGAACUGAGUGCAGCACUCGGAAAA--A-AAUUGGGAACAACCAUUUGAAUUUG (((-----(.......(((((((((.((((((....(((((((.....)))))))...))..)))).)))....))))))...--.-...(((......))).))))..... ( -29.50) >DroSim_CAF1 4779 109 + 1 UCAGUUCAGCGAGAACUCCGAAGCAGUCAGCUAUGAACAUGUCCUUGAGACAUGUAAGAGAACUGAGUGCAGCACUCGGAAAA--A-AUUUGGGAACAACCAUUUGAAUUUG ..(((((((.......(((((.(((.((((((....(((((((.....)))))))...))..)))).))).....)))))...--.-...(((......))).))))))).. ( -28.70) >DroEre_CAF1 12831 102 + 1 UCAGUUCAGCGAGAACUCCGAGGCAGUCAGCUGUGAACAUGUCCUUGAGACAUGUA-GAGAACUGAGUGC---ACUCAGAGAA--A-AAUUGGUAGCAACUGCUA---UCAG ...............(((.((((((.((((.(.(..(((((((.....))))))).-.).).)))).)))---.))).)))..--.-...(((((((....))))---))). ( -35.40) >DroYak_CAF1 5891 110 + 1 UCAGUUCAGCGAGAAGUCCGAGGCAGUCAGCUGUGAACAUGUCCUUGAGACAUGUAAGAGAAUUCAGUGCAACACUCAAAAAA--ACUAUUGAAAGCGACUUUUAGAAUUGG .((((((....(((((((....((((....))))..(((((((.....)))))))....(..(((((((..............--..)))))))..)))))))).)))))). ( -26.19) >consensus UCAGUUCAGCGAGAACUCCGAGGCAGUCAGCUAUGAACAUGUCCUUGAGACAUGUAAGAGAACUGAGUGCAGCACUCGGAAAA__A_AAUUGGGAACAACCAUUUGAAUUUG ................(((((((((.((((......(((((((.....))))))).......)))).)))....))))))..........(((......))).......... (-17.58 = -18.78 + 1.20)

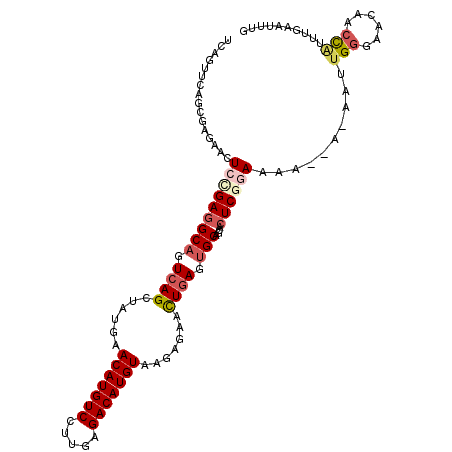
| Location | 11,747,466 – 11,747,571 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11747466 105 - 22407834 ------CAAAUGGUUUUUCCCAACU-UUUUUUUUCGAGUGCCGCUCUCAGUUCUCUUACAUGUCUCAAGGACAUGUUCAUAGCUGACUGCCUCAGAGUUCUCGCUGAACUGA ------....(((......)))...-.........(((....((..((((((.....(((((((.....)))))))....))))))..)))))..(((((.....))))).. ( -26.00) >DroSec_CAF1 5919 104 - 1 CAAAUUCAAAUGGUUGUUCCCAAUU-U--UUUUCCGAGUGCUGCACUCAGUUCUCUUACAUGUCUCAAGGACAUGUUCAUAGCUGACUGCCUCGGAGUUCUCAC-----UGA ..........(((......)))...-.--..(((((((....(((.((((((.....(((((((.....)))))))....)))))).)))))))))).......-----... ( -29.70) >DroSim_CAF1 4779 109 - 1 CAAAUUCAAAUGGUUGUUCCCAAAU-U--UUUUCCGAGUGCUGCACUCAGUUCUCUUACAUGUCUCAAGGACAUGUUCAUAGCUGACUGCUUCGGAGUUCUCGCUGAACUGA ..........(((......)))...-.--...((((((....(((.((((((.....(((((((.....)))))))....)))))).)))))))))((((.....))))... ( -30.60) >DroEre_CAF1 12831 102 - 1 CUGA---UAGCAGUUGCUACCAAUU-U--UUCUCUGAGU---GCACUCAGUUCUC-UACAUGUCUCAAGGACAUGUUCACAGCUGACUGCCUCGGAGUUCUCGCUGAACUGA .((.---((((....)))).))...-.--...((((((.---(((.((((((...-.(((((((.....)))))))....)))))).)))))))))((((.....))))... ( -36.80) >DroYak_CAF1 5891 110 - 1 CCAAUUCUAAAAGUCGCUUUCAAUAGU--UUUUUUGAGUGUUGCACUGAAUUCUCUUACAUGUCUCAAGGACAUGUUCACAGCUGACUGCCUCGGACUUCUCGCUGAACUGA .((.(((...((((((((......)))--.....((((.((..(((((.........(((((((.....)))))))...))).))...)))))))))))......))).)). ( -20.30) >consensus CAAAUUCAAAUGGUUGUUCCCAAUU_U__UUUUCCGAGUGCUGCACUCAGUUCUCUUACAUGUCUCAAGGACAUGUUCAUAGCUGACUGCCUCGGAGUUCUCGCUGAACUGA ...............................(((((((....(((.((((((.....(((((((.....)))))))....)))))).))))))))))............... (-22.46 = -22.78 + 0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:21 2006