
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,737,825 – 11,737,940 |
| Length | 115 |
| Max. P | 0.854458 |

| Location | 11,737,825 – 11,737,940 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -24.58 |
| Energy contribution | -25.82 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11737825 115 + 22407834 AUGUAUCAUGUGCUCCGUUAAUCAAACGUUGCCAUACCAACAGGAGCACGUGACUCACGAUAAGACCGGCGUACUUAGGACGGUUUCCAUAGAACGCG-----CCUGAUAAACAAAGCAC .((..((((((((((((((.....)))((((......)))).)))))))))))..))..........(((((.((..(((.....)))..))...)))-----))............... ( -34.00) >DroSim_CAF1 197448 114 + 1 AUGAAUCAUGUGCUCCGUUAAUCAAACGUUGCCAUACCAACAGGAGCACGUGACUCACGAUAAGACCGGCGUACUUAGGACGGUUUCCAUAGAACGCG-----C-UGAUAAACAAAGCAC .(((.((((((((((((((.....)))((((......)))).))))))))))).)))...........((((.((..(((.....)))..)).))))(-----(-(.........))).. ( -37.00) >DroYak_CAF1 191148 115 + 1 AUGAAUCAUGUGCUCCGUUAAUCAAACGUUGCCAUACCAACCGGAGCACGUGACUCACGAUAAGACCGGCGUACUUAGGACGGUUUCCAUAGAACGCG-----CCUGAUAAACAAAGCAC .(((.((((((((((((((.....)))((((......)))).))))))))))).)))..........(((((.((..(((.....)))..))...)))-----))............... ( -34.80) >DroAna_CAF1 194979 119 + 1 AUGAAUCAUGUGCUCCAGCAAUCAUUCGUUGCCAUAUCAACUAGGGCACGUGACUCUCGAUAAGACCUUCGUACUUAAGAUGGU-ACCAUAGAGCCCGUACUGCUUGAUAAACAAAGCAC .....((((((((((..(((((.....)))))...........))))))))))..(((......(((.((........)).)))-......))).......(((((........))))). ( -27.02) >consensus AUGAAUCAUGUGCUCCGUUAAUCAAACGUUGCCAUACCAACAGGAGCACGUGACUCACGAUAAGACCGGCGUACUUAGGACGGUUUCCAUAGAACGCG_____CCUGAUAAACAAAGCAC ..((.((((((((((((((.....)))((((......)))).))))))))))).))............((((.((..(((.....)))..)).))))....................... (-24.58 = -25.82 + 1.25)

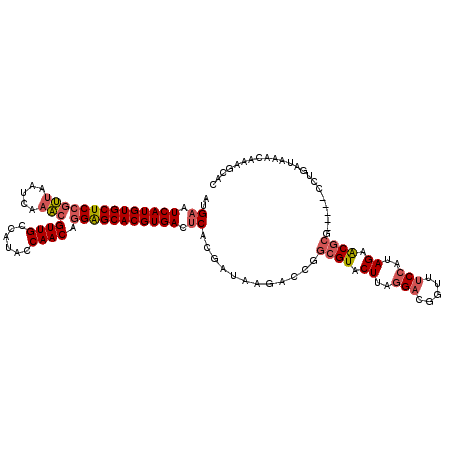
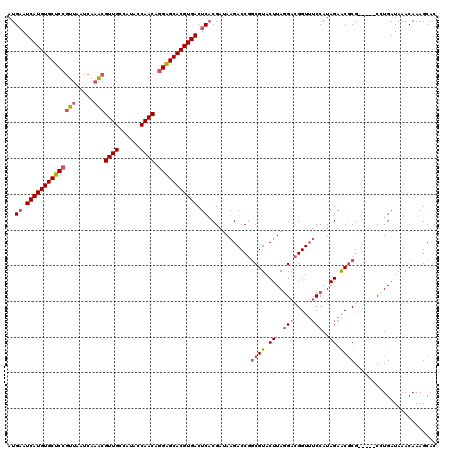
| Location | 11,737,825 – 11,737,940 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.79 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -27.94 |
| Energy contribution | -28.25 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11737825 115 - 22407834 GUGCUUUGUUUAUCAGG-----CGCGUUCUAUGGAAACCGUCCUAAGUACGCCGGUCUUAUCGUGAGUCACGUGCUCCUGUUGGUAUGGCAACGUUUGAUUAACGGAGCACAUGAUACAU ((((((((((.((((((-----(((((.((..(((.....)))..)).))))..(((.(((((.((((.....))))....))))).)))...))))))).))))))))))......... ( -39.20) >DroSim_CAF1 197448 114 - 1 GUGCUUUGUUUAUCA-G-----CGCGUUCUAUGGAAACCGUCCUAAGUACGCCGGUCUUAUCGUGAGUCACGUGCUCCUGUUGGUAUGGCAACGUUUGAUUAACGGAGCACAUGAUUCAU ..(((.........)-)-----)((((.((..(((.....)))..)).))))..........((((((((.(((((((.(((((((((....)))...))))))))))))).)))))))) ( -38.30) >DroYak_CAF1 191148 115 - 1 GUGCUUUGUUUAUCAGG-----CGCGUUCUAUGGAAACCGUCCUAAGUACGCCGGUCUUAUCGUGAGUCACGUGCUCCGGUUGGUAUGGCAACGUUUGAUUAACGGAGCACAUGAUUCAU ((((((((((.((((((-----((.((.((..(((.....)))..)).))((((..((.((((.((((.....)))))))).))..))))..)))))))).))))))))))......... ( -42.30) >DroAna_CAF1 194979 119 - 1 GUGCUUUGUUUAUCAAGCAGUACGGGCUCUAUGGU-ACCAUCUUAAGUACGAAGGUCUUAUCGAGAGUCACGUGCCCUAGUUGAUAUGGCAACGAAUGAUUGCUGGAGCACAUGAUUCAU (((((((((..((((..(.(((((((((((.((((-(..(((((.......)))))..))))))))))).)))))....((((......)))))..)))).)).)))))))......... ( -36.40) >consensus GUGCUUUGUUUAUCAGG_____CGCGUUCUAUGGAAACCGUCCUAAGUACGCCGGUCUUAUCGUGAGUCACGUGCUCCUGUUGGUAUGGCAACGUUUGAUUAACGGAGCACAUGAUUCAU ((((((((.....))...............((((...))))...))))))............((((((((.(((((((.(((((((((....)))...))))))))))))).)))))))) (-27.94 = -28.25 + 0.31)

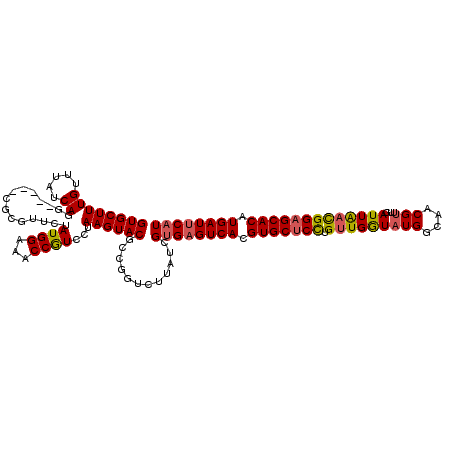
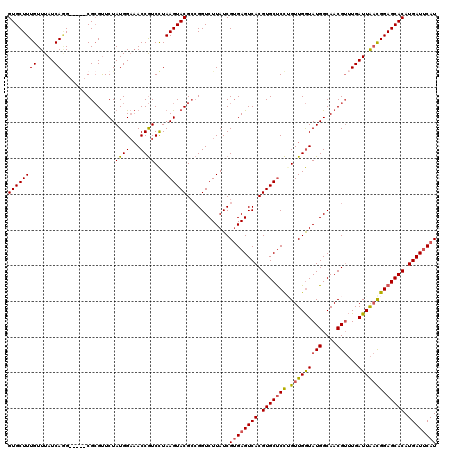
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:14 2006