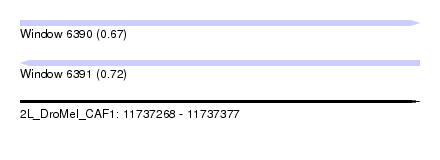
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,737,268 – 11,737,377 |
| Length | 109 |
| Max. P | 0.715462 |

| Location | 11,737,268 – 11,737,377 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 113 |
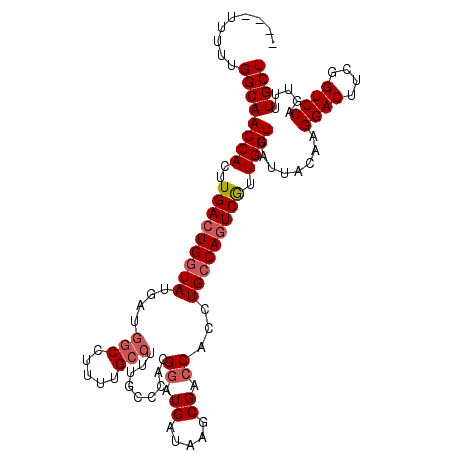
| Reading direction | forward |
| Mean pairwise identity | 92.49 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -26.45 |
| Energy contribution | -27.07 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11737268 109 + 22407834 AGCAAAACUGGACCGAAGUCCUUGUAAUGACCAACGACUGGCAGGUGGUCGCUUAUCAUCCGGGGGCAAAGGCAAAAGGCCAUCAUAGCAGUCAAGUGGUUGCCAAAAA---- .((((.(((.(((....(((((((..((((....(((((.......)))))....)))).)))))))...(((.....))).........))).)))..))))......---- ( -30.50) >DroSec_CAF1 197120 109 + 1 AGCAAAACUGGACCGAAGUCCUUGUAAUGACCAAUGACUGGCAGGUGGUCGCUUAUCAUUCGUGGGCAAAGGCAAAAGGCCAUCAUACCAGUCAAGUGGUUGCCAAAAA---- .((...((.((((....))))..))...(((((.(((((((...(((((.((((((.....))))))...(((.....)))))))).)))))))..)))))))......---- ( -35.40) >DroSim_CAF1 196913 109 + 1 AGCAAAACUGGACCGAAGUCCUUGUAAUGACCAACGACUGGCAGGUGGUCGCUUAUCAUCCGUGGGCAAAGGCAGAAGGCCAUUAUACCAGUCAAGUGGUUGCCAAAAA---- .((...((.((((....))))..))...(((((..((((((...(((((.((((((.....))))))...(((.....)))))))).))))))...)))))))......---- ( -32.40) >DroYak_CAF1 190626 110 + 1 AGCAAAACUGGACCGAAGUCCUUGUAAUGACCAACGACUGGCAGGUGGUCGCUUAUCAUCCGCGGG--AUGCCAAAAGGCGAUCAUACCA-UCAAGUGGUUGCCAAAAAAUAU .((...((.((((....))))..))...(((((..((.(((...((((((((((..(((((...))--))).....)))))))))).)))-))...))))))).......... ( -32.60) >consensus AGCAAAACUGGACCGAAGUCCUUGUAAUGACCAACGACUGGCAGGUGGUCGCUUAUCAUCCGUGGGCAAAGGCAAAAGGCCAUCAUACCAGUCAAGUGGUUGCCAAAAA____ .((...((.((((....))))..))...(((((..((((((..(((((((.........(....)((....))....)))))))...))))))...))))))).......... (-26.45 = -27.07 + 0.62)

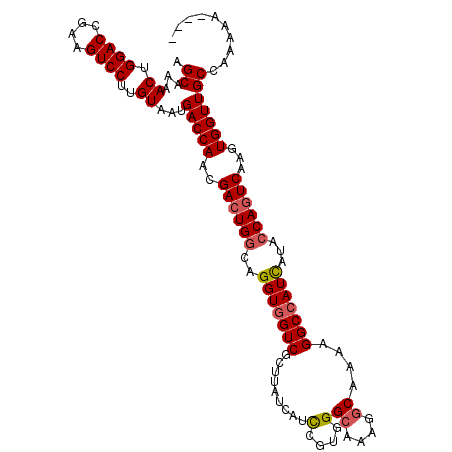
| Location | 11,737,268 – 11,737,377 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 92.49 |
| Mean single sequence MFE | -32.97 |
| Consensus MFE | -27.41 |
| Energy contribution | -28.48 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11737268 109 - 22407834 ----UUUUUGGCAACCACUUGACUGCUAUGAUGGCCUUUUGCCUUUGCCCCCGGAUGAUAAGCGACCACCUGCCAGUCGUUGGUCAUUACAAGGACUUCGGUCCAGUUUUGCU ----.....(((((..(((.(((((.......(((.....))).......((.((((((.((((((.........)))))).))))))....))....))))).))).))))) ( -29.00) >DroSec_CAF1 197120 109 - 1 ----UUUUUGGCAACCACUUGACUGGUAUGAUGGCCUUUUGCCUUUGCCCACGAAUGAUAAGCGACCACCUGCCAGUCAUUGGUCAUUACAAGGACUUCGGUCCAGUUUUGCU ----.....((((((((..(((((((((.(.(((((((...(.((((....)))).)..))).).)))).))))))))).))))........((((....)))).....)))) ( -36.20) >DroSim_CAF1 196913 109 - 1 ----UUUUUGGCAACCACUUGACUGGUAUAAUGGCCUUCUGCCUUUGCCCACGGAUGAUAAGCGACCACCUGCCAGUCGUUGGUCAUUACAAGGACUUCGGUCCAGUUUUGCU ----.....((((((((..(((((((((....(((.....))).........((.((.....)).))...))))))))).))))........((((....)))).....)))) ( -33.90) >DroYak_CAF1 190626 110 - 1 AUAUUUUUUGGCAACCACUUGA-UGGUAUGAUCGCCUUUUGGCAU--CCCGCGGAUGAUAAGCGACCACCUGCCAGUCGUUGGUCAUUACAAGGACUUCGGUCCAGUUUUGCU .........((((((((..(((-(((((((.((((.((....(((--((...)))))..)))))).))..))))).))).))))........((((....)))).....)))) ( -32.80) >consensus ____UUUUUGGCAACCACUUGACUGGUAUGAUGGCCUUUUGCCUUUGCCCACGGAUGAUAAGCGACCACCUGCCAGUCGUUGGUCAUUACAAGGACUUCGGUCCAGUUUUGCU .........((((((((..(((((((((....(((.....))).........((.((.....)).))...))))))))).))))........((((....)))).....)))) (-27.41 = -28.48 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:12 2006