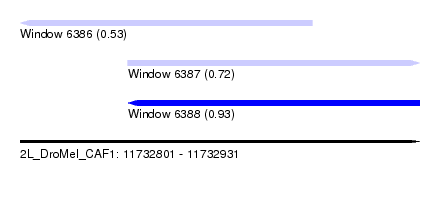
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,732,801 – 11,732,931 |
| Length | 130 |
| Max. P | 0.929349 |

| Location | 11,732,801 – 11,732,896 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -11.84 |
| Energy contribution | -11.95 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.41 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
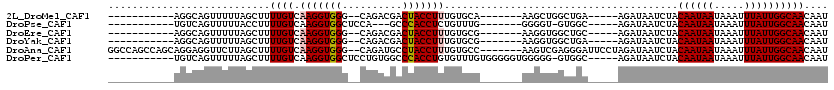
>2L_DroMel_CAF1 11732801 95 - 22407834 -----------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCA-------AAGCUGGCUGA-----AGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU -----------..((.(((((((((((((((((((((((.--.......))))).))).)))-------))...))))))-----))))......((((((.....))))))))...... ( -27.00) >DroPse_CAF1 187417 93 - 1 -----------UGUCAGUUUUUACCUUUUGUCAAGGUGGCUCCA---GCCCACCUCUGUUUG-------GGGGU-GUGGC-----AGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU -----------((((.....(((((((.....))))))).....---((((((((((....)-------)))))-).)))-----.)))).....((((((.....))))))........ ( -27.20) >DroEre_CAF1 210695 95 - 1 -----------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCG-------AAGGUGGCUGC-----AGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU -----------..((((((....((((((((((((((((.--.......))))).))).)))-------)))))))))))-----..........((((((.....))))))........ ( -27.00) >DroYak_CAF1 185101 95 - 1 -----------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCG-------AAGGUGGCUGA-----AGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU -----------..((.(((((((((((((((((((((((.--.......))))).))).)))-------))...))))))-----))))......((((((.....))))))))...... ( -26.70) >DroAna_CAF1 190373 111 - 1 GGCCAGCCAGCAGGAGGUUCUUAGCUUUUGUCAAGGUGGG--CAGAUGCCUACCUUUGUGCC-------AAGUCGAGGGAUUCCUAGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU ((....)).(((((((.((((..((((....(((((((((--(....))))))).)))....-------))))..)))).)))))..........((((((.....))))))))...... ( -35.60) >DroPer_CAF1 184022 103 - 1 -----------UGUCAGUUUUUAGCUUUUGUCAAGGUGGCUCCUGUGGCCCACCUGUGUUUGUGGGGGUGGGGG-GUGGC-----AGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU -----------(((((((.((((....((((..((((.....((((.((((.((..(.((....)).)..))))-)).))-----))...))))))))...))))...)))))))..... ( -30.10) >consensus ___________AGGCAGUUUUUAGCUUUUGUCAAGGUGGG__CAGACGACUACCUUUGUGCG_______AAGGUGGCGGA_____AGAUAAUCUACAAUAAUAAAUUUAUUGGCAACAAU ...........................((((.(((((((..........))))))).......................................((((((.....)))))))))).... (-11.84 = -11.95 + 0.11)

| Location | 11,732,836 – 11,732,931 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -15.15 |
| Energy contribution | -14.82 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11732836 95 + 22407834 UCAGCCAGCUUUGCACAAAGGUAGUCGUCUG--CCCACCUUGACAAAAGCUAAAAACUGCCU--------------GCCGGAUGCCGAGUUAACCCUUGGCAU--CCAACAAU ......((((((...(((.(((((....)))--))....)))...))))))...........--------------...((((((((((......))))))))--))...... ( -30.10) >DroPse_CAF1 187452 95 + 1 GCCAC-ACCCCCAAACAGAGGUGGGC---UGGAGCCACCUUGACAAAAGGUAAAAACUGACA--------------ACCGGAUGCCGGGUUAAACCCUGGCCUGGCCAACAAU (((.(-(((.(......).)))))))---(((....(((((.....)))))...........--------------.((((..((((((......)))))))))))))..... ( -31.60) >DroSim_CAF1 185014 95 + 1 UCAGCCACCUUCGCACAAAGGUAGUCGUCUG--CCCACCUUGACAAAAGCUAAAAACUGCCU--------------GCCGGAUGCCGAGUUAACCCUUGGCAU--CCAACAAU ...((.......)).....(((((..(((..--........)))....((........))))--------------)))((((((((((......))))))))--))...... ( -28.30) >DroEre_CAF1 210730 95 + 1 GCAGCCACCUUCGCACAAAGGUAGUCGUCUG--CCCACCUUGACAAAAGCUAAAAACUGCCU--------------GCCGGAUGCCGAGUUAACCCUUGGCAU--CCAACAAU ((((.((((((......))))).((((....--.......))))..............).))--------------)).((((((((((......))))))))--))...... ( -29.30) >DroYak_CAF1 185136 95 + 1 UCAGCCACCUUCGCACAAAGGUAGUCGUCUG--CCCACCUUGACAAAAGCUAAAAACUGCCU--------------GCCGGAUGCCGAGUUAACCCUUGGCAU--CCAACAAU ...((.......)).....(((((..(((..--........)))....((........))))--------------)))((((((((((......))))))))--))...... ( -28.30) >DroAna_CAF1 190413 109 + 1 UCCCUCGACUUGGCACAAAGGUAGGCAUCUG--CCCACCUUGACAAAAGCUAAGAACCUCCUGCUGGCUGGCCGAUGCCGGAUGCCGGGUUAACCCUUGGCAU--CCAACAAU ........((((((...(((((.(((....)--)).))))).......))))))...........(((........)))((((((((((....)))..)))))--))...... ( -39.80) >consensus UCAGCCACCUUCGCACAAAGGUAGUCGUCUG__CCCACCUUGACAAAAGCUAAAAACUGCCU______________GCCGGAUGCCGAGUUAACCCUUGGCAU__CCAACAAU .................(((((.(..........).)))))......................................((((((((((......))))))))..))...... (-15.15 = -14.82 + -0.33)

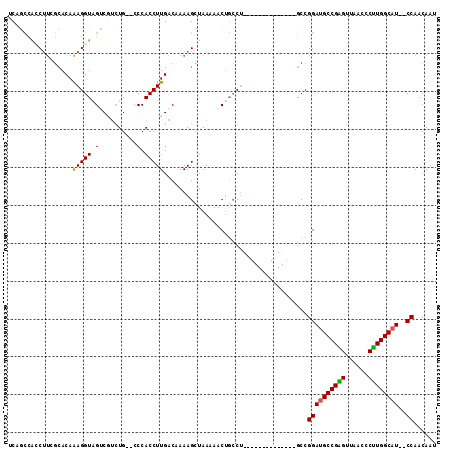
| Location | 11,732,836 – 11,732,931 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -23.33 |
| Energy contribution | -23.37 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
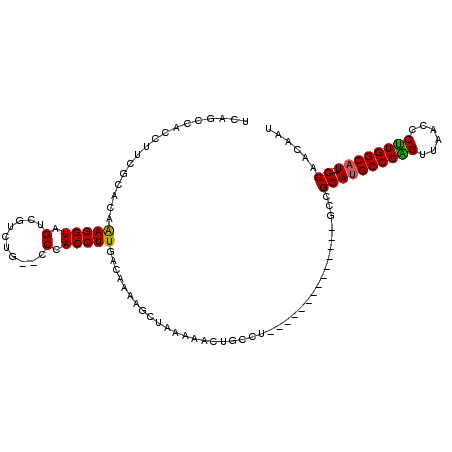
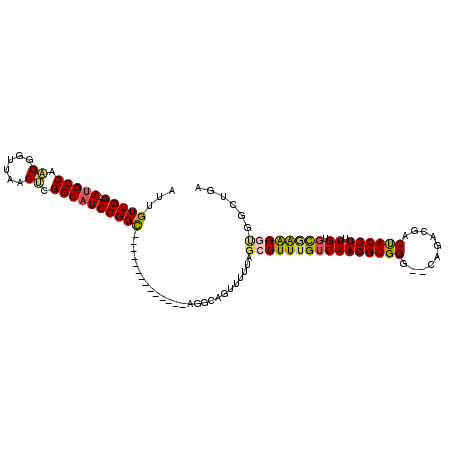
>2L_DroMel_CAF1 11732836 95 - 22407834 AUUGUUGG--AUGCCAAGGGUUAACUCGGCAUCCGGC--------------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCAAAGCUGGCUGA .(((((((--(((((.((......)).))))))))))--------------)).((((...(((((((.((((((((((.--.......))))).))).))))))))))))). ( -40.70) >DroPse_CAF1 187452 95 - 1 AUUGUUGGCCAGGCCAGGGUUUAACCCGGCAUCCGGU--------------UGUCAGUUUUUACCUUUUGUCAAGGUGGCUCCA---GCCCACCUCUGUUUGGGGGU-GUGGC .......((((.(((.(((.....))))))....(((--------------((..(((...((((((.....))))))))).))---)))(((((((....))))))-))))) ( -34.80) >DroSim_CAF1 185014 95 - 1 AUUGUUGG--AUGCCAAGGGUUAACUCGGCAUCCGGC--------------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCGAAGGUGGCUGA .(((((((--(((((.((......)).))))))))))--------------)).(((((....((((((((((((((((.--.......))))).))).))))))))))))). ( -40.90) >DroEre_CAF1 210730 95 - 1 AUUGUUGG--AUGCCAAGGGUUAACUCGGCAUCCGGC--------------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCGAAGGUGGCUGC .(((((((--(((((.((......)).))))))))))--------------))((((((....((((((((((((((((.--.......))))).))).)))))))))))))) ( -43.00) >DroYak_CAF1 185136 95 - 1 AUUGUUGG--AUGCCAAGGGUUAACUCGGCAUCCGGC--------------AGGCAGUUUUUAGCUUUUGUCAAGGUGGG--CAGACGACUACCUUUGUGCGAAGGUGGCUGA .(((((((--(((((.((......)).))))))))))--------------)).(((((....((((((((((((((((.--.......))))).))).))))))))))))). ( -40.90) >DroAna_CAF1 190413 109 - 1 AUUGUUGG--AUGCCAAGGGUUAACCCGGCAUCCGGCAUCGGCCAGCCAGCAGGAGGUUCUUAGCUUUUGUCAAGGUGGG--CAGAUGCCUACCUUUGUGCCAAGUCGAGGGA ......((--(((((..(((....))))))))))((((((.(((.(((.(((((((.(....).)))))))...))).))--).))))))..((((((........)))))). ( -50.30) >consensus AUUGUUGG__AUGCCAAGGGUUAACUCGGCAUCCGGC______________AGGCAGUUUUUAGCUUUUGUCAAGGUGGG__CAGACGACUACCUUUGUGCGAAGGUGGCUGA ...(((((..(((((.((......)).))))))))))..........................((((((((((((((((..........)))))).)).))))))))...... (-23.33 = -23.37 + 0.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:09 2006