
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,732,572 – 11,732,726 |
| Length | 154 |
| Max. P | 0.996848 |

| Location | 11,732,572 – 11,732,663 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -20.93 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.99 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11732572 91 + 22407834 GCAGAGGCAUCCUUUUCCCACGCCUUACAGCACGCUCUUACCAAAACACAUGCAUACAAACACAAAGGCGUCGGAAAAUGGA-----------GAAAAGCCA .....(((.(((((((((.(((((((..((....))..............((....))......))))))).)))))).)))-----------.....))). ( -23.50) >DroSec_CAF1 192518 89 + 1 GCAGAGGCAUCCUUUUCCCACGCCU-------------UACCAACACACAUACAUACAAACACAAAGGCGUCGGAAAAUGGAAAUGGAAAAUGGAAAAGCCA .....(((.(((((((((.((((((-------------(.........................))))))).)))))).)))..(....)........))). ( -23.01) >DroSim_CAF1 184760 78 + 1 GCAGAGGCAUCCUUUUCCCACGCCU-------------UACCAACACACACACAUACAAACACAAAGGCGUCGGAAAAUGGA-----------GAAAAGCCA .....(((.(((((((((.((((((-------------(.........................))))))).)))))).)))-----------.....))). ( -23.61) >DroEre_CAF1 210470 89 + 1 GCAGAGGCAUCCUUUUCCUACGACUUUCAGUACAUUCUUGCCAGUACAC--ACACACAAACACAAAGGCGACGGAAAAUGGA-----------GAAAAGCCA .....(((.((((((((((((........))).....(((((.......--...............))))).)))))).)))-----------.....))). ( -18.15) >DroYak_CAF1 184873 89 + 1 GCACUGGUAUCCUUUUCCUACGACUUUCGGCACGUUCUUGUCCAUACAC--AAAUACAAACAUAAAGGCGACGGAAAAUGGA-----------GAAAAGCCA .....(((.(((((((((..((.((((......((..((((......))--))..))......)))).))..)))))).)))-----------.....))). ( -16.40) >consensus GCAGAGGCAUCCUUUUCCCACGCCUU_C_G_AC__UCUUACCAACACACA_ACAUACAAACACAAAGGCGUCGGAAAAUGGA___________GAAAAGCCA .....(((.(((((((((.(((((((......................................))))))).)))))).)))................))). (-14.95 = -15.99 + 1.04)

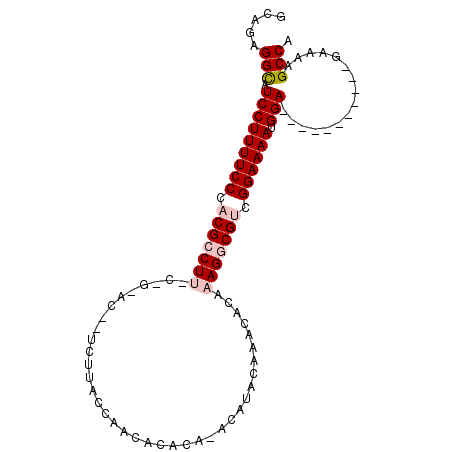

| Location | 11,732,572 – 11,732,663 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.78 |
| Mean single sequence MFE | -25.45 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.81 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11732572 91 - 22407834 UGGCUUUUC-----------UCCAUUUUCCGACGCCUUUGUGUUUGUAUGCAUGUGUUUUGGUAAGAGCGUGCUGUAAGGCGUGGGAAAAGGAUGCCUCUGC .(((.....-----------(((.((((((.(((((((...........((((((.(((....))).))))))...))))))).))))))))).)))..... ( -32.24) >DroSec_CAF1 192518 89 - 1 UGGCUUUUCCAUUUUCCAUUUCCAUUUUCCGACGCCUUUGUGUUUGUAUGUAUGUGUGUUGGUA-------------AGGCGUGGGAAAAGGAUGCCUCUGC .(((...(((.((((((((..((.....(((((((...((..(....)..))...)))))))..-------------.)).)))))))).))).)))..... ( -28.00) >DroSim_CAF1 184760 78 - 1 UGGCUUUUC-----------UCCAUUUUCCGACGCCUUUGUGUUUGUAUGUGUGUGUGUUGGUA-------------AGGCGUGGGAAAAGGAUGCCUCUGC .(((.....-----------(((.((((((.(((((((......(..((....))..).....)-------------)))))).))))))))).)))..... ( -25.30) >DroEre_CAF1 210470 89 - 1 UGGCUUUUC-----------UCCAUUUUCCGUCGCCUUUGUGUUUGUGUGU--GUGUACUGGCAAGAAUGUACUGAAAGUCGUAGGAAAAGGAUGCCUCUGC .(((.....-----------(((.((((((..((.((((......(((..(--.(((....)))...)..)))..)))).))..))))))))).)))..... ( -20.00) >DroYak_CAF1 184873 89 - 1 UGGCUUUUC-----------UCCAUUUUCCGUCGCCUUUAUGUUUGUAUUU--GUGUAUGGACAAGAACGUGCCGAAAGUCGUAGGAAAAGGAUACCAGUGC (((((((((-----------((.((((((.(.(((.(((.((((..(((..--..)))..)))).))).)))).)))))).)..)))))))....))).... ( -21.70) >consensus UGGCUUUUC___________UCCAUUUUCCGACGCCUUUGUGUUUGUAUGU_UGUGUAUUGGUAAGA__GU_C_G_AAGGCGUGGGAAAAGGAUGCCUCUGC .(((................(((.((((((.(((((((..(((..((((......))))..)))............))))))).))))))))).)))..... (-16.09 = -16.81 + 0.72)

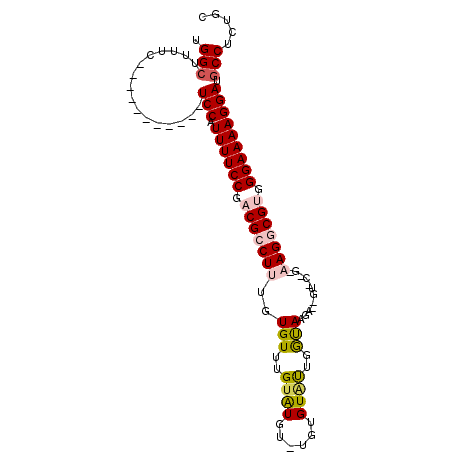
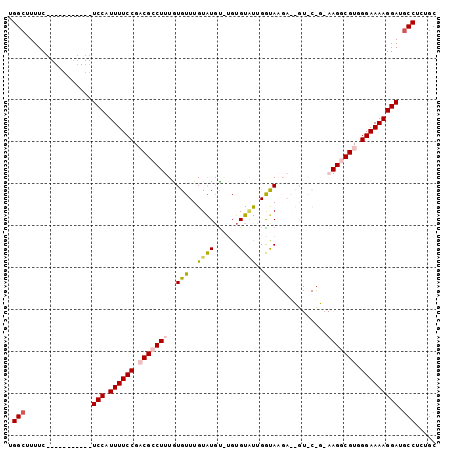
| Location | 11,732,634 – 11,732,726 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -20.95 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11732634 92 + 22407834 CAAAGGCGUCGGAAAAUGGAGAAAAGCCAAGCUUUGAGCUCAGUUCUGGCUACAAAAA--AUGCAGGGAAAAAAAGGUAAAGCAUCAAAGCUGG ....(((.((..........))...))).(((((((((((...((((.((........--..))..))))..........))).)))))))).. ( -20.32) >DroEre_CAF1 210530 90 + 1 CAAAGGCGACGGAAAAUGGAGAAAAGCCAGGCUUCGAGCUGAGUUCUUUCAACAAAAA--AUGUAGGGA-A-AAAGGUAAAGCAUCAAAGCUGG ....(((..................))).(((((.(((((...((((((((.......--.)).)))))-)-........))).)).))))).. ( -15.37) >DroYak_CAF1 184933 93 + 1 UAAAGGCGACGGAAAAUGGAGAAAAGCCAGGCUUUGAGCUGAGUCCUUGCAACAACAAGGAUGUUGCGAAA-AAAGGUAAAGCAUCAAAGCUGG ....(((..................))).(((((((((((....((((((((((.......))))))....-.))))...))).)))))))).. ( -27.17) >consensus CAAAGGCGACGGAAAAUGGAGAAAAGCCAGGCUUUGAGCUGAGUUCUUGCAACAAAAA__AUGUAGGGAAA_AAAGGUAAAGCAUCAAAGCUGG ....(((..................))).(((((((((((....((((((((((.......))))))......))))...))).)))))))).. (-18.52 = -19.30 + 0.79)

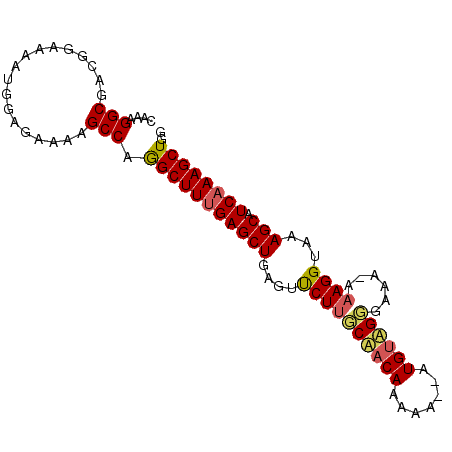
| Location | 11,732,634 – 11,732,726 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.81 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -15.01 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11732634 92 - 22407834 CCAGCUUUGAUGCUUUACCUUUUUUUCCCUGCAU--UUUUUGUAGCCAGAACUGAGCUCAAAGCUUGGCUUUUCUCCAUUUUCCGACGCCUUUG ..((((((((.((((.......((((..(((((.--....)))))..))))..)))))))))))).(((..................))).... ( -19.57) >DroEre_CAF1 210530 90 - 1 CCAGCUUUGAUGCUUUACCUUU-U-UCCCUACAU--UUUUUGUUGAAAGAACUCAGCUCGAAGCCUGGCUUUUCUCCAUUUUCCGUCGCCUUUG ((((((((((.(((.....(((-(-((...(((.--....))).))))))....))))))))).)))).......................... ( -18.30) >DroYak_CAF1 184933 93 - 1 CCAGCUUUGAUGCUUUACCUUU-UUUCGCAACAUCCUUGUUGUUGCAAGGACUCAGCUCAAAGCCUGGCUUUUCUCCAUUUUCCGUCGCCUUUA ((((((((((.(((...((((.-....((((((.......))))))))))....))))))))).)))).......................... ( -24.00) >consensus CCAGCUUUGAUGCUUUACCUUU_UUUCCCUACAU__UUUUUGUUGCAAGAACUCAGCUCAAAGCCUGGCUUUUCUCCAUUUUCCGUCGCCUUUG ((((((((((.(((.....(((......(((((.......)))))...)))...)))))))))).))).......................... (-15.01 = -14.47 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:06 2006