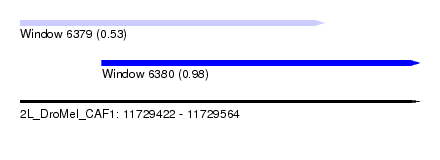
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,729,422 – 11,729,564 |
| Length | 142 |
| Max. P | 0.979266 |

| Location | 11,729,422 – 11,729,530 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.57 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -16.84 |
| Energy contribution | -17.96 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11729422 108 + 22407834 GUAUCUUUAACAAAUGAUAUCGCAUUACUUGAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCUACAUUCUGUGCAUCCAGAAUGCUAGAA (((((..........)))))..................(((((..(((((.....)))))(((.....)))))))).((((((((((((......))))))).))))) ( -29.50) >DroSec_CAF1 188840 108 + 1 GUACCUUUAACAAAUUAUAUCGCAUUACUUCAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCCACAUUCUGUGCAUCCAGAAUGCUAGAA ...................((.................(((((..(((((.....)))))(((.....))))))))......(((((((......)))))))...)). ( -24.60) >DroSim_CAF1 181077 108 + 1 GUACCUUUAACAAAUUAUAUCGCAUUACUUCAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCCACAUUUUGUGCAUCGAGAAUGCUAGAA .....................(((((.(((........(((((..(((((.....)))))(((.....))))))))....(((.....)))....))))))))..... ( -24.60) >DroEre_CAF1 206703 108 + 1 GUACCUUUAACAAAUAAUAUCGCAUUAUUUGAACAGUUAUACUAGCCAGCAAUAUGCUGAGGCUUUGAGCCAGUAUGUUUAACAUUUUGAACAUGCAGUGAGAUCGAA ..........((((((((.....))))))))..(.(((.((((...((((.....)))).(((.....))).((((((((((....)))))))))))))).))).).. ( -26.30) >DroYak_CAF1 181150 108 + 1 GUACCUUUAACAAAUUAUUUCGCAUUACUUGAACAUUAAUACUCGCCAGCAAUAUGCUGAGGCUUUGUGCCUGUAUGUUCUGCAUUCUGUGCAAGGAAAAUGCUUGAA .....................(((((....((((((..........((((.....))))((((.....))))..))))))(((((...))))).....)))))..... ( -24.20) >consensus GUACCUUUAACAAAUUAUAUCGCAUUACUUGAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCCACAUUCUGUGCAUCCAGAAUGCUAGAA ...................((.........(((((....((((..(((((.....)))))(((.....))))))))))))..(((((((......)))))))...)). (-16.84 = -17.96 + 1.12)

| Location | 11,729,451 – 11,729,564 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -31.24 |
| Consensus MFE | -19.40 |
| Energy contribution | -20.76 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11729451 113 + 22407834 UGAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCUACAUUCUGUGCAUCCAGAAUGCUAGAAUGUCCUAAAUUCCCACAGCCAAUGCUUGAAUUAA------ .(((((....((((..(((((.....)))))(((.....)))))))))))).((((((((.((((.....))))))))))))....(((((....(((....))).)))))..------ ( -33.30) >DroSec_CAF1 188869 119 + 1 UCAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCCACAUUCUGUGCAUCCAGAAUGCUAGAAUGUACUAAAUUCCCACAGCCAAUGCUUGAAUUAAAUGCUC ((((((((.(((((..(((((.....)))))(((.....)))))))).....((((((((.((((.....)))))))))))).................)))).))))........... ( -33.00) >DroSim_CAF1 181106 119 + 1 UCAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCCACAUUUUGUGCAUCGAGAAUGCUAGAAUGUACUAAAUUCCCACAGCCAAUGCUUGGAUUAAAUGCUC ....((((.(((((..(((((.....)))))(((.....)))))))).....((((((((.((((.....)))))))))))).........(((.(((....))))))....))))... ( -31.40) >DroEre_CAF1 206732 110 + 1 UGAACAGUUAUACUAGCCAGCAAUAUGCUGAGGCUUUGAGCCAGUAUGUUUAACAUUUUGAACAUGCAGUGAGAUCGAAUGGUCCAAAUUCGCACAGCCAAUUGUUCAUU--------- (((((((((........((((.....)))).((((........((((((((((....)))))))))).(((....(((((.......)))))))))))))))))))))..--------- ( -34.20) >DroYak_CAF1 181179 110 + 1 UGAACAUUAAUACUCGCCAGCAAUAUGCUGAGGCUUUGUGCCUGUAUGUUCUGCAUUCUGUGCAAGGAAAAUGCUUGAAUUUCCCAAAUACCCACAGCCAGCGCUCGAAU--------- .((((((..........((((.....))))((((.....))))..)))))).((...(((((...((.((((......)))).)).......)))))...))........--------- ( -24.30) >consensus UGAACAUUAAUACCAACCAGCAAUAUGUUGGGGCUUUGUGCCGGUAUGUUCCACAUUCUGUGCAUCCAGAAUGCUAGAAUGUACUAAAUUCCCACAGCCAAUGCUUGAAUUAA______ .(((((....((((..(((((.....)))))(((.....)))))))))))).((((((((.((((.....))))))))))))..................................... (-19.40 = -20.76 + 1.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:02 2006