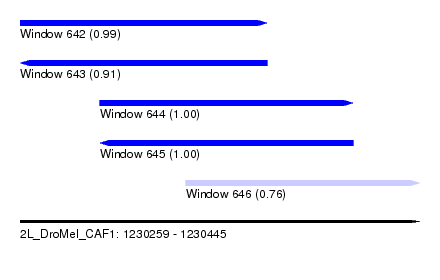
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,230,259 – 1,230,445 |
| Length | 186 |
| Max. P | 0.999967 |

| Location | 1,230,259 – 1,230,374 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -33.24 |
| Consensus MFE | -27.26 |
| Energy contribution | -27.02 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1230259 115 + 22407834 UGCCGCCGGAGG---GAAAUUUAAGUGCCUUCGGCGACCAAACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGGAAAA--AAAAAUGCUCAACAAAAU ...(((((((((---.(........).))))))))).............(((((((((((....((((..(((((....)))))...........)))).--.....))))..))))))) ( -31.22) >DroSec_CAF1 7614 113 + 1 UGCCGCCGGAGG---GAAAUUUAAGUGCCCUCGGCGACCAAACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAACAAAAU ...(((((.(((---(...........))))))))).............(((((((.(.(((........(((((....)))))......(((...--..--...))))))).))))))) ( -32.10) >DroSim_CAF1 3410 113 + 1 UGCCGCCGGAGG---GAAAUUUAAGUGCCUUCGGCGACCAAACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAGCAAAAU ...(((((((((---.(........).))))))))).............(((((((.(.(((........(((((....)))))......(((...--..--...))))))).))))))) ( -34.30) >DroEre_CAF1 3573 113 + 1 UGUCGCCGGAGG---GAAAUUUAAGUGCCUCUGGCGACCAAGCGAACCAAUUUUGAAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAACAAAAU .(((((((((((---.(........).)))))))))))...........((((((..(.(((........(((((....)))))......(((...--..--...)))))))..)))))) ( -37.10) >DroYak_CAF1 3445 118 + 1 UGGCGCUGGGGGGCGGAAAUUUAAGUGCCUUCGGCGACCAAACGAACCAAUUUUGAAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGCCCGA--AAAAAAAGAUGCCCAACAAAAU ((((((((((((((..........)).))))))))).))).........((((((..(.((((......)(((((....)))))............--..........))))..)))))) ( -31.50) >consensus UGCCGCCGGAGG___GAAAUUUAAGUGCCUUCGGCGACCAAACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA__AA__AAAGAUGCCCAACAAAAU ...(((((((((...............))))))))).............(((((((.(.((((......)(((((....)))))........................)))).))))))) (-27.26 = -27.02 + -0.24)

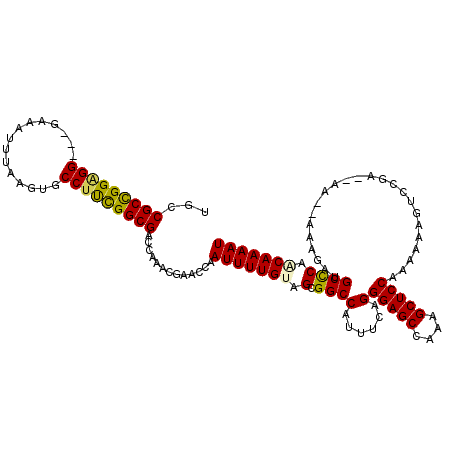
| Location | 1,230,259 – 1,230,374 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -35.68 |
| Consensus MFE | -28.47 |
| Energy contribution | -28.87 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1230259 115 - 22407834 AUUUUGUUGAGCAUUUUU--UUUUCCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUUUGGUCGCCGAAGGCACUUAAAUUUC---CCUCCGGCGGCA ........((((...(((--(.(..(((...((((.(((((....)))))..))))...)))..).))))...)))).....(((((((.(((............---))).))))))). ( -35.00) >DroSec_CAF1 7614 113 - 1 AUUUUGUUGGGCAUCUUU--UU--UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUUUGGUCGCCGAGGGCACUUAAAUUUC---CCUCCGGCGGCA (((((((.((((......--((--((((........(((((....)))))))))))..))).).)))))))...........(((((((((((...........)---))).))))))). ( -38.40) >DroSim_CAF1 3410 113 - 1 AUUUUGCUGGGCAUCUUU--UU--UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUUUGGUCGCCGAAGGCACUUAAAUUUC---CCUCCGGCGGCA ((((((..((((......--((--((((........(((((....)))))))))))..))).)..))))))...........(((((((.(((............---))).))))))). ( -34.10) >DroEre_CAF1 3573 113 - 1 AUUUUGUUGGGCAUCUUU--UU--UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUUCAAAAUUGGUUCGCUUGGUCGCCAGAGGCACUUAAAUUUC---CCUCCGGCGACA ..................--..--.((((((.(((((((((...((((.........)))))))))))))..))))))....((((((.((((............---)))).)))))). ( -36.40) >DroYak_CAF1 3445 118 - 1 AUUUUGUUGGGCAUCUUUUUUU--UCGGGCUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUUCAAAAUUGGUUCGUUUGGUCGCCGAAGGCACUUAAAUUUCCGCCCCCCAGCGCCA .....((((((...........--..((((..(((((((((...((((.........)))))))))))))..((...(((((((.(((...))))).)))))..)))))))))))).... ( -34.51) >consensus AUUUUGUUGGGCAUCUUU__UU__UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUUUGGUCGCCGAAGGCACUUAAAUUUC___CCUCCGGCGGCA ........((((.............(((.(((((..(((((....)))))..)))).).)))...........)))).....(((((((.(((...............))).))))))). (-28.47 = -28.87 + 0.40)

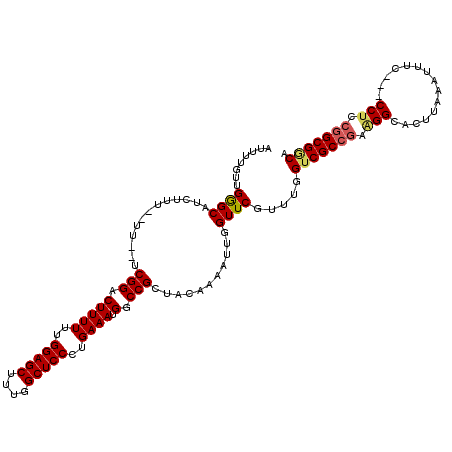
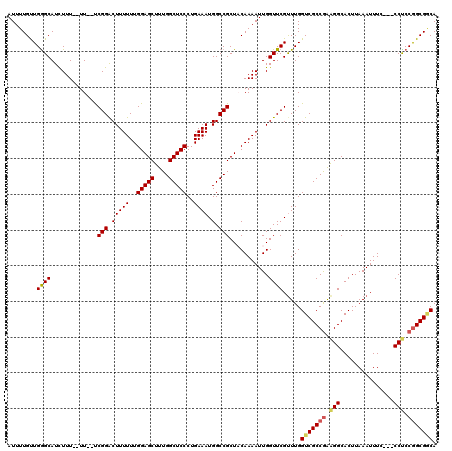
| Location | 1,230,296 – 1,230,414 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.34 |
| Mean single sequence MFE | -33.83 |
| Consensus MFE | -30.92 |
| Energy contribution | -30.96 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.99 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1230296 118 + 22407834 AACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGGAAAA--AAAAAUGCUCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCA .(((((.....)))))...((((.......(((((....)))))......((.((((...--........(((......))).(((((((.......)))))))....)))).)))))). ( -36.20) >DroSec_CAF1 7651 116 + 1 AACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCA .(((((.....)))))...((((....((((((((....))))).....((((...--..--.........((......))..(((((((.......))))))).)))))))...)))). ( -34.00) >DroSim_CAF1 3447 116 + 1 AACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAGCAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCA .(((((.....)))))...((((....((((((((....))))).....((((...--..--.....(((...))).......(((((((.......))))))).)))))))...)))). ( -36.00) >DroEre_CAF1 3610 112 + 1 AGCGAACCAAUUUUGAAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAACAAAAUGAAGCGGUAUGAAGUUAAUGCCGCAGACU----ACGGCCA .((..............))((((.......(((((....))))).....((((...--..--.........((......))..(((((((.......))))))).))))----..)))). ( -31.44) >DroYak_CAF1 3485 114 + 1 AACGAACCAAUUUUGAAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGCCCGA--AAAAAAAGAUGCCCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACU----GCGGCCA ...................((((....((((((((....)))))............--.........................(((((((.......)))))))...))----).)))). ( -31.50) >consensus AACGAACCAAUUUUGUAGCGGCCAUUUCAGGGAGCCAAAGCUCCAAAAAAGUCCGA__AA__AAAGAUGCCCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCA ...................((((.......(((((....))))).....((((..................((......))..(((((((.......))))))).))))......)))). (-30.92 = -30.96 + 0.04)

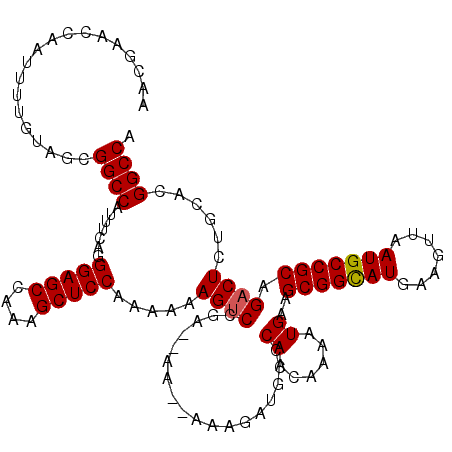
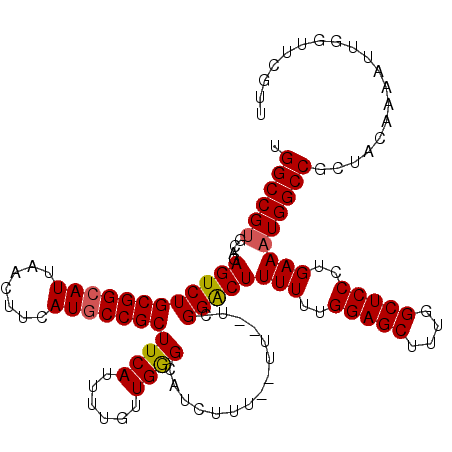
| Location | 1,230,296 – 1,230,414 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.34 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -32.96 |
| Energy contribution | -33.04 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1230296 118 - 22407834 UGGCCGUGCAGAGUCUGCGGCAUUAACUUCAUGCCGCUUCAUUUUGUUGAGCAUUUUU--UUUUCCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUU .((((((.(((((((((((((((.......)))))))((((......)))).......--......))))).....(((((....))))))))..))))))((((......))))..... ( -38.20) >DroSec_CAF1 7651 116 - 1 UGGCCGUGCAGAGUCUGCGGCAUUAACUUCAUGCCGCUUCAUUUUGUUGGGCAUCUUU--UU--UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUU ..(((..(((((((..(((((((.......)))))))...)))))))..)))......--..--.((((((.(((((.(((...((((.........))))))).)))))..)))))).. ( -38.50) >DroSim_CAF1 3447 116 - 1 UGGCCGUGCAGAGUCUGCGGCAUUAACUUCAUGCCGCUUCAUUUUGCUGGGCAUCUUU--UU--UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUU ((.(((.(((((((..(((((((.......)))))))...)))))))))).)).....--..--.((((((.(((((.(((...((((.........))))))).)))))..)))))).. ( -40.20) >DroEre_CAF1 3610 112 - 1 UGGCCGU----AGUCUGCGGCAUUAACUUCAUACCGCUUCAUUUUGUUGGGCAUCUUU--UU--UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUUCAAAAUUGGUUCGCU ..(((((----.....)))))..............(((.((......)))))......--..--.((((((.(((((((((...((((.........)))))))))))))..)))))).. ( -30.80) >DroYak_CAF1 3485 114 - 1 UGGCCGC----AGUCUGCGGCAUUAACUUCAUGCCGCUUCAUUUUGUUGGGCAUCUUUUUUU--UCGGGCUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUUCAAAAUUGGUUCGUU ((.((((----((...(((((((.......)))))))......))).))).)).........--.((((((.(((((((((...((((.........)))))))))))))..)))))).. ( -36.40) >consensus UGGCCGUGCAGAGUCUGCGGCAUUAACUUCAUGCCGCUUCAUUUUGUUGGGCAUCUUU__UU__UCGGACUUUUUUGGAGCUUUGGCUCCCUGAAAUGGCCGCUACAAAAUUGGUUCGUU .((((((....((((((((((((.......)))))))((((......))))...............)))))(((..(((((....)))))..)))))))))................... (-32.96 = -33.04 + 0.08)

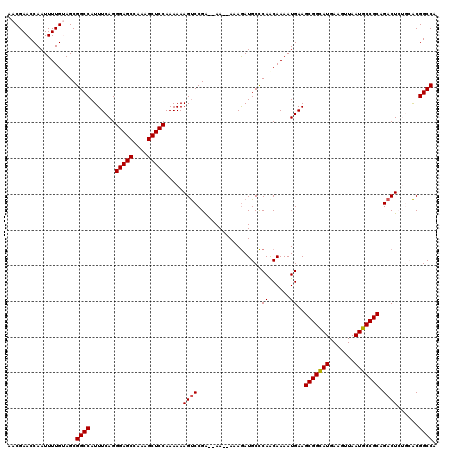
| Location | 1,230,336 – 1,230,445 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.34 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -22.26 |
| Energy contribution | -21.98 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1230336 109 + 22407834 CUCCAAAAAAGUCCGGAAAA--AAAAAUGCUCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCAUGUGCUU---------CACGAGACAGCGGAUGCCAAAAAG ..........((.((((...--........(((......))).(((((((.......)))))))....)))).))(((.((.(((((---------(....)).)))).)))))...... ( -28.40) >DroSec_CAF1 7691 107 + 1 CUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCAUGUGCUC---------CACGAGACAGCGGAUGCCAAAAAG ..........(((((.--..--.....................(((((((.......)))))))...(((.(((((....)))))..---------...)))....)))))......... ( -27.00) >DroSim_CAF1 3487 107 + 1 CUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAGCAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCAUGUGCUC---------CACGAGACAGCGGAUGCCAAAAAG ..........(((((.--..--......(((..(((.......(((((((.......)))))))......)))..)))..(((.(((---------...)))))).)))))......... ( -27.12) >DroEre_CAF1 3650 103 + 1 CUCCAAAAAAGUCCGA--AA--AAAGAUGCCCAACAAAAUGAAGCGGUAUGAAGUUAAUGCCGCAGACU----ACGGCCAUGUGCUC---------CACGAGACAGCGGAUGCCAAAAAG ..........(((((.--..--......(((((......))..(((((((.......))))))).....----..)))..(((.(((---------...)))))).)))))......... ( -22.50) >DroYak_CAF1 3525 114 + 1 CUCCAAAAAAGCCCGA--AAAAAAAGAUGCCCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACU----GCGGCCAUGUGCUCCAUGUGCUUCACGAGACAGCGGAUGCCAAAAAG ..........(((((.--.........((.....))...(((((((.((((.(((....(((((.....----))))).....))).)))))))))))........)))..))....... ( -28.10) >consensus CUCCAAAAAAGUCCGA__AA__AAAGAUGCCCAACAAAAUGAAGCGGCAUGAAGUUAAUGCCGCAGACUCUGCACGGCCAUGUGCUC_________CACGAGACAGCGGAUGCCAAAAAG ..........(((((.............(((((......))..(((((((.......)))))))...........)))..(((.(((............)))))).)))))......... (-22.26 = -21.98 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:12 2006