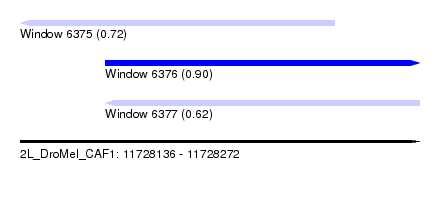
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,728,136 – 11,728,272 |
| Length | 136 |
| Max. P | 0.902876 |

| Location | 11,728,136 – 11,728,243 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 88.07 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -17.08 |
| Energy contribution | -18.12 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11728136 107 - 22407834 CCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUACAGCCACACACCCACUUCCACACCCAUAUUAGAGAUGCG ....((((.((..((((.....))))..))(((..(((..(((((.....)))))..)))..)))....................................).))). ( -22.70) >DroSec_CAF1 187557 105 - 1 CCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGGCACACACCCACAACCACACCCAUAUUAG--AUGCG ....((((.((..((((.....))))..))..((((((((((((((.........)))))....)))))))))..........................)--.))). ( -27.60) >DroSim_CAF1 179804 105 - 1 CCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGGCACACACCCACAUCCACACCCAUAUUAG--AUGCG ....((((.((..((((.....))))..))..((((((((((((((.........)))))....)))))))))..........................)--.))). ( -27.60) >DroEre_CAF1 205445 96 - 1 CCAAGCACAGAUGCUUUUCAAAGAAGUGGGAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGCCAC--ACCCACAUC-------GUAAUAG--AUGCG ..(((((....)))))......((.(((((.(((.(((((((((((.........)))))....)))))))))..--.))))).))-------(((....--.))). ( -24.50) >DroYak_CAF1 179838 97 - 1 CC------AGCUGCUUUUCAAAGGAGUGCCAGGCCUGAAAAUGACUCCCAGUCAUAGUCACUCCUUUUCAGUCAC--ACCCACAUCCACACCCUUAGUAG--AUGCG ..------.((((......((((((((((((....))...(((((.....))))).).))))))))).))))...--.....((((.((.......)).)--))).. ( -19.90) >consensus CCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGCCACACACCCACAUCCACACCCAUAUUAG__AUGCG ....(((..((((((((.....))))))))(((..(((..(((((.....)))))..)))..)))......................................))). (-17.08 = -18.12 + 1.04)


| Location | 11,728,165 – 11,728,272 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -34.84 |
| Consensus MFE | -27.64 |
| Energy contribution | -28.16 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11728165 107 + 22407834 UGUGUGGCUGUAAAGGAGUGACUAUGACUAGGAGUCAUUUUCAGGCCUGCCACUUCUUUGAAAAGCAGCUGUGCUUGGCUGAAAGCCGCAGUUGAUUUGCAAAGCCC ...(((((.....(((..(((..(((((.....)))))..)))..))))))))..(((((.....(((((((((((......))).)))))))).....)))))... ( -38.00) >DroSec_CAF1 187584 107 + 1 UGUGUGCCUGAAAAGGAGUGACUAUGACUAGGAGUCAUUUUCAGGCCUGCCACUUCUUUGAAAAGCAGCUGUGCUUGGCUCAAAGCCGCAGUUGAUUUGCAAAGCCC .(((.((((((....((((((((.........))))))))))))))....)))..(((((.....(((((((((((......))).)))))))).....)))))... ( -35.30) >DroSim_CAF1 179831 107 + 1 UGUGUGCCUGAAAAGGAGUGACUAUGACUAGGAGUCAUUUUCAGGCCUGCCACUUCUUUGAAAAGCAGCUGUGCUUGGCUCAAAGCCGCAGUUGAUUUGCAAAGCCC .(((.((((((....((((((((.........))))))))))))))....)))..(((((.....(((((((((((......))).)))))))).....)))))... ( -35.30) >DroEre_CAF1 205465 105 + 1 U--GUGGCUGAAAAGGAGUGACUAUGACUAGGAGUCAUUUUCAGGCCUCCCACUUCUUUGAAAAGCAUCUGUGCUUGGCUCAAAGCCGCAGUUGAUUUGCAAAGCCC .--..((((..((((((((((..(((((.....)))))..)).((....))))))))))...(((((....)))))(((.....)))((((.....))))..)))). ( -30.90) >DroYak_CAF1 179865 99 + 1 U--GUGACUGAAAAGGAGUGACUAUGACUGGGAGUCAUUUUCAGGCCUGGCACUCCUUUGAAAAGCAGCU------GGUUCAAAGCCGCUGCUGAUUUGCAAAGCCC (--(..(....(((((((((.(((.(.(((..((...))..))).).))))))))))))....((((((.------(((.....)))))))))...)..))...... ( -34.70) >consensus UGUGUGCCUGAAAAGGAGUGACUAUGACUAGGAGUCAUUUUCAGGCCUGCCACUUCUUUGAAAAGCAGCUGUGCUUGGCUCAAAGCCGCAGUUGAUUUGCAAAGCCC ...(((.......(((..(((..(((((.....)))))..)))..)))..)))..(((((.....(((((((....(((.....)))))))))).....)))))... (-27.64 = -28.16 + 0.52)


| Location | 11,728,165 – 11,728,272 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 92.13 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -23.14 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11728165 107 - 22407834 GGGCUUUGCAAAUCAACUGCGGCUUUCAGCCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUACAGCCACACA .((((..(((.......)))(((.....))).......((..((((.....))))..))(((..(((..(((((.....)))))..)))..)))....))))..... ( -31.20) >DroSec_CAF1 187584 107 - 1 GGGCUUUGCAAAUCAACUGCGGCUUUGAGCCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGGCACACA ..((((.(((.......)))(((.....)))))))...((..((((.....))))..))..((((((((((((((.........)))))....)))))))))..... ( -34.50) >DroSim_CAF1 179831 107 - 1 GGGCUUUGCAAAUCAACUGCGGCUUUGAGCCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGGCACACA ..((((.(((.......)))(((.....)))))))...((..((((.....))))..))..((((((((((((((.........)))))....)))))))))..... ( -34.50) >DroEre_CAF1 205465 105 - 1 GGGCUUUGCAAAUCAACUGCGGCUUUGAGCCAAGCACAGAUGCUUUUCAAAGAAGUGGGAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGCCAC--A .((((...........(..(..(((((((..(((((....))))))))))))..)..)((((..(((..(((((.....)))))..)))..))))...))))..--. ( -32.70) >DroYak_CAF1 179865 99 - 1 GGGCUUUGCAAAUCAGCAGCGGCUUUGAACC------AGCUGCUUUUCAAAGGAGUGCCAGGCCUGAAAAUGACUCCCAGUCAUAGUCACUCCUUUUCAGUCAC--A ...(((((.(((..((((((((.......))------.))))))))))))))((.((..(((..(((..(((((.....)))))..)))..)))...)).))..--. ( -28.60) >consensus GGGCUUUGCAAAUCAACUGCGGCUUUGAGCCAAGCACAGCUGCUUUUCAAAGAAGUGGCAGGCCUGAAAAUGACUCCUAGUCAUAGUCACUCCUUUUCAGCCACACA ((((((.(((.......)))(((.....))).......((((((((.....))))))))))))))((..(((((.....)))))..))................... (-23.14 = -23.98 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:59 2006