
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,724,382 – 11,724,473 |
| Length | 91 |
| Max. P | 0.984504 |

| Location | 11,724,382 – 11,724,473 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.41 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -15.40 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11724382 91 + 22407834 AGUUGCCGCAAACAGAGUCUGGCGCAAAAGAUCAAUGACUCCAUUGAAGUUCAAUAA-AAUCGUCCUCGACCACGCUAGACUUUUCCA--------AAA-------A--- .(((......)))(((((((((((.......((((((....))))))..........-..(((....)))...)))))))))))....--------...-------.--- ( -22.90) >DroPse_CAF1 172767 110 + 1 AUUUGCCGUAAACAAAGUCUGGCACAAAAGAUCAAUGACUCCAUUGAAGUUCCAUAAAAAUUGUCCAACGCCAUCCCAGACUUCCACACACAAAAAAAAAAAAAAAGACG .((((..((.....((((((((.........((((((....)))))).(((..(((.....)))..)))......))))))))....)).))))................ ( -16.30) >DroEre_CAF1 201622 91 + 1 AGUUGCCGCAAACAGAGUCUGGCGCAAAAGAUCAAUGACUCCAUUGAAGUUCAAUAA-AAUUGUCCUCGGCCACGCUAGACUUCCCCA--------AAA-------A--- .(((......))).((((((((((.......((((((....))))))((..((((..-.))))..))......)))))))))).....--------...-------.--- ( -21.90) >DroYak_CAF1 175713 91 + 1 AGUUGCCGCAAACAGAGUCUGGCGCAAAAGAUCAAUGACUCCAUUGAAGUUCAAUAA-AAUCGUCCUCGGCCACGCUAGACUUCCCCA--------AAA-------A--- .(((......))).((((((((((.......((((((....))))))..........-..(((....)))...)))))))))).....--------...-------.--- ( -21.40) >DroAna_CAF1 181587 86 + 1 ------------AAGAGUCUGACUCAAAAGAUCAAUGACUUUGUUGAAGUUCAAUAA-AAUUGUCCUCGGUCACGGUAGACUUUACCA--------GAAUAAAAAAA--- ------------.((((((((.((.....((((...((((((((((.....))))))-)...)))...))))..))))))))))....--------...........--- ( -16.50) >DroPer_CAF1 176488 105 + 1 AUUUGCCGUAAACAAAGUCUGGCACAAAAGAUCAAUGACUCCAUUGAAGUUCCAUAAAAAUUGUCCAACGCCAUCCCAGACUUCCACACCA-----AAAAAAAAAAAACG .((((..((.....((((((((.........((((((....)))))).(((..(((.....)))..)))......))))))))..))..))-----))............ ( -15.70) >consensus AGUUGCCGCAAACAGAGUCUGGCGCAAAAGAUCAAUGACUCCAUUGAAGUUCAAUAA_AAUUGUCCUCGGCCACGCUAGACUUCCCCA________AAA_______A___ ..............((((((((((.......((((((....))))))...............((.....))..))))))))))........................... (-15.40 = -14.73 + -0.66)

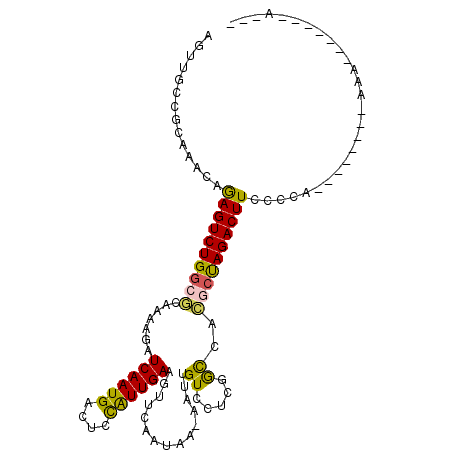

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:56 2006