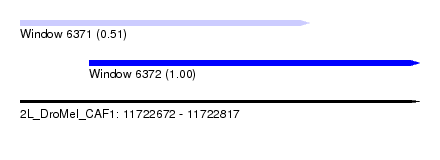
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,722,672 – 11,722,817 |
| Length | 145 |
| Max. P | 0.998757 |

| Location | 11,722,672 – 11,722,777 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -21.04 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
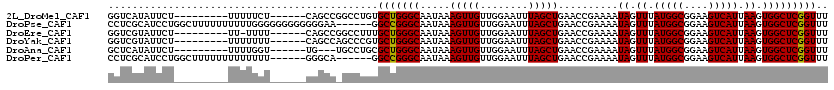
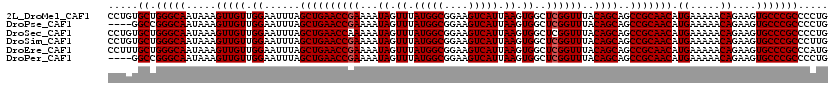
>2L_DroMel_CAF1 11722672 105 + 22407834 GGUCAUAUUCU---------UUUUUCU------CAGCCGGCCUGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU (((...((((.---------.......------...(((((....))))).(((((....)))))))))...)))(((((((...((.((.(((((....))))).)).))..))))))) ( -28.50) >DroPse_CAF1 171129 114 + 1 CCUCGCAUCCUGGCUUUUUUUUUUGGGGGGGGGGGGAA------GGCCGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU ....((..((((((((((((((((.....)))))))))------)))))))(((((....))))).......)).(((((((...((.((.(((((....))))).)).))..))))))) ( -37.30) >DroEre_CAF1 199949 104 + 1 GGUCGUAUUCU---------UU-UUUU------CAGCCGGCCUUUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU (((...((((.---------..-....------...(((((....))))).(((((....)))))))))...)))(((((((...((.((.(((((....))))).)).))..))))))) ( -28.50) >DroYak_CAF1 173967 105 + 1 GGUCGUAUUCU---------UUUUUUU------CAGCCAGCCCGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU (((...((((.---------.......------...(((((....))))).(((((....)))))))))...)))(((((((...((.((.(((((....))))).)).))..))))))) ( -29.20) >DroAna_CAF1 179966 102 + 1 GCUCAUAUUCU---------UUUUGGU------UG---UGCCUGCGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU (((...(((((---------....(.(------(.---(((((.....)))))...)).)....)))))..))).(((((((...((.((.(((((....))))).)).))..))))))) ( -30.20) >DroPer_CAF1 174916 108 + 1 CCUCGCAUCCUGGCUUUUUUUUUUUUU------GGGCA------GGCCGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU ....((..((((((((.(((.......------))).)------)))))))(((((....))))).......)).(((((((...((.((.(((((....))))).)).))..))))))) ( -31.60) >consensus GCUCGUAUUCU_________UUUUUUU______CAGCC_GCCUGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUU .............................................(((((((.....(((((........)))))..........((.((.(((((....))))).)).))))))))).. (-21.04 = -20.82 + -0.22)
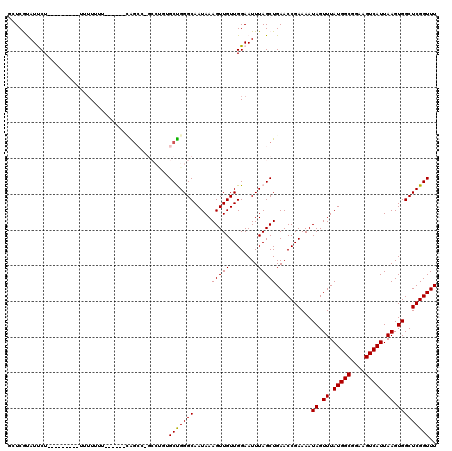
| Location | 11,722,697 – 11,722,817 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.38 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -35.53 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
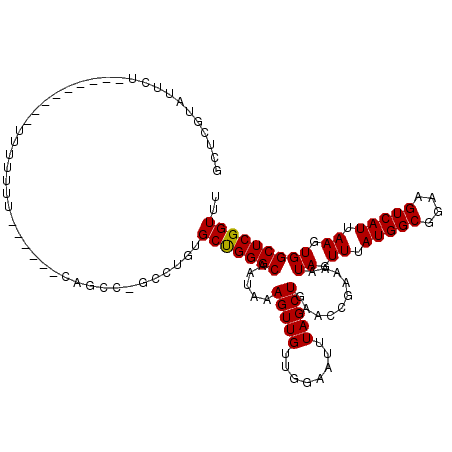
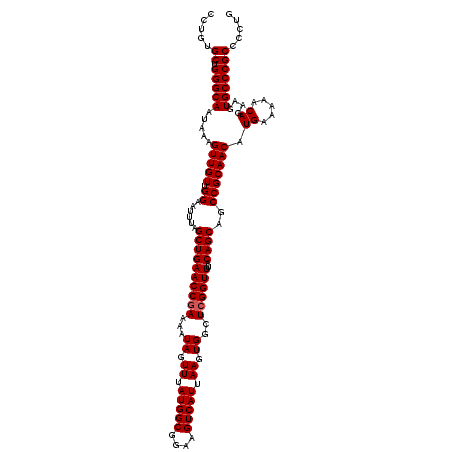
>2L_DroMel_CAF1 11722697 120 + 22407834 CCUGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCCUG .....((.(((((.....(((((.((......((((((((((...((.((.(((((....))))).)).))..))))))..))))..))))))).((.....))....)))))))..... ( -36.50) >DroPse_CAF1 171167 116 + 1 ----GGCCGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCCUG ----(((.(((((.....(((((.((......((((((((((...((.((.(((((....))))).)).))..))))))..))))..))))))).((.....))....)))))))).... ( -39.80) >DroSec_CAF1 182089 120 + 1 CCUGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCAAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCCUG ........((((......(((((((......((((((.(((..........(((((....)))))....))).))))))..))))))).(((...((.....))....)))..))))... ( -33.34) >DroSim_CAF1 174308 120 + 1 CCUGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCUUG .....((.(((((.....(((((.((......((((((((((...((.((.(((((....))))).)).))..))))))..))))..))))))).((.....))....)))))))..... ( -36.50) >DroEre_CAF1 199973 120 + 1 CCUUUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCAUG .....((.(((((.....(((((.((......((((((((((...((.((.(((((....))))).)).))..))))))..))))..))))))).((.....))....)))))))..... ( -36.50) >DroPer_CAF1 174948 116 + 1 ----GGCCGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCCUG ----(((.(((((.....(((((.((......((((((((((...((.((.(((((....))))).)).))..))))))..))))..))))))).((.....))....)))))))).... ( -39.80) >consensus CCUGUGCUGGGCAAUAAAGUUGUUGGAAUUUAGCUGAACCGAAAAUAGUUUAUGGCGGAAGUCAUUAAGUGGCUCGGUUUACAGCAGCCGCAACAUGAAAAACAGAAGUGCCCGCCCCUG .....((.(((((.....(((((.((......((((((((((...((.((.(((((....))))).)).))..))))))..))))..))))))).((.....))....)))))))..... (-35.53 = -35.70 + 0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:54 2006