| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,707,823 – 11,707,927 |
| Length | 104 |
| Max. P | 0.844015 |

| Location | 11,707,823 – 11,707,927 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.19 |
| Mean single sequence MFE | -29.35 |
| Consensus MFE | -24.53 |
| Energy contribution | -24.50 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
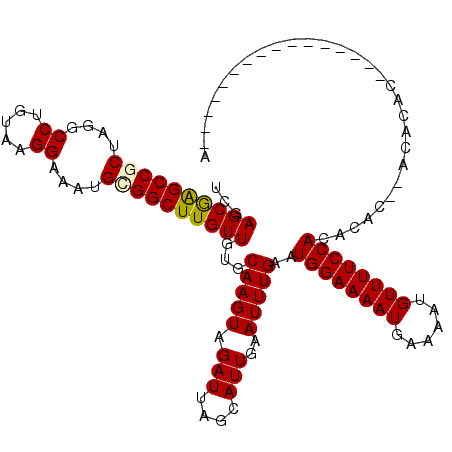
>2L_DroMel_CAF1 11707823 104 - 22407834 UCGAGCGAGCCGCUAGGCCUGUAAGGAAAUGUGGCUUGUUGUGCAAGUAGAUUAGCAUUGAAUUUGAAUGGAAAAUGAAAAUGUUUUCCACACACACACACAC----------------A ...((((((((((....((.....))....))))))))))((((((((.(((....)))..)))))..((((((((......)))))))))))..........----------------. ( -28.50) >DroPse_CAF1 161135 93 - 1 UCGAGCGAGCCUCUCGGCC----GGGAAAUGCGGCUUGUUCGGCAAGUAGAUUAGCAUUGGAUUUGAAUGGAAAAUGAAAAUGUUUUCCACACCC--AG--------------------- (((((((((((((((....----)))).....)))))))))))(((((.(((....)))..)))))..((((((((......)))))))).....--..--------------------- ( -27.60) >DroEre_CAF1 192177 105 - 1 UCGAGCGAGCCGCUAGGCCUGUAAGGAAAUGUGGCUUGUUGUGCAAGUGGAUUAGCAUUGAAUUUGAAUGGAAAAUGAAAAUGUUUUCCACACAC--ACACAC----------AC---UA ...((((((((((....((.....))....))))))))))(((...(((((..((((((..((((.......))))...)))))).)))))...)--))....----------..---.. ( -33.40) >DroYak_CAF1 166279 108 - 1 UCGAGCGAGCCGCUAGGCCUGUAAGGAAAUGUGGCUUGUUGUGCAAGUAGAUUAGCAUUGAAUUUGAAUGGAAAAUGAAAAUGUUUUCCACACAC--ACAUAC----------ACUCUUA ..(((((((((((....((.....))....)))))))).(((((((((.(((....)))..)))))..((((((((......))))))))....)--)))...----------.)))... ( -28.60) >DroAna_CAF1 172954 112 - 1 UUGAGCAGGCCGCU---CCUUGCAGGAAAUGCGGCUUGUUGUGCAAGUAGAUUAGCAUUGAAUUUGAAUGGAAAAUGAAAAUGUUUUCCACUCUC--ACACAGACACACACAGAC---CA ...(((((((((((---((.....)))...))))))))))((((((((......)).))).....((.((((((((......)))))))).))..--.)))..............---.. ( -30.70) >DroPer_CAF1 162235 93 - 1 CCGAGCGAGCCUCUCGGCC----GGGAAAUGCGGCUUGUUCUGCAAGUAGAUUAGCAUUGGAUUUGAAUGGAAAAUGAAAAUGUUUUCCACACCC--AG--------------------- (((((.......)))))..----(((.(((((.(((((.....)))))......))))).........((((((((......))))))))..)))--..--------------------- ( -27.30) >consensus UCGAGCGAGCCGCUAGGCCUGUAAGGAAAUGCGGCUUGUUGUGCAAGUAGAUUAGCAUUGAAUUUGAAUGGAAAAUGAAAAUGUUUUCCACACAC__ACACAC________________A ...((((((((((....((.....))....))))))))))...(((((.(((....)))..)))))..((((((((......)))))))).............................. (-24.53 = -24.50 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:49 2006