| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,703,720 – 11,703,813 |
| Length | 93 |
| Max. P | 0.838572 |

| Location | 11,703,720 – 11,703,813 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
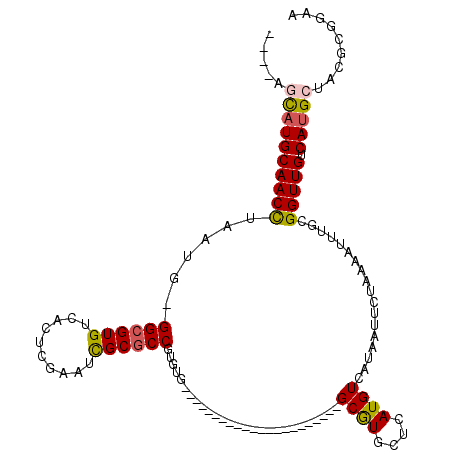
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.28 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11703720 93 + 22407834 ----AGCAUGCAACCUAAUG-GGCGUGUCACUCGAAUCGCGCCGUGUG----------------------GCGUGCUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGCUACGCGGAA ----(((((((((((..(((-((((((((((.((........)).)))----------------------))))))))))((...((((....)))).))))))).))))))........ ( -33.40) >DroSec_CAF1 170485 93 + 1 ----AGCAUGCAACCUAAUG-GGCGUGUCACUCGAAUUGCGCCGUGUG----------------------GCAUGCUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGCUACGCGGAA ----(((((((((((..(((-((((((((((.((........)).)))----------------------))))))))))((...((((....)))).))))))).))))))........ ( -36.80) >DroEre_CAF1 188037 93 + 1 ----GGCAUGCAACCUAAUG-GGCGUGUCACUCGAAUCGCGCCGUGUG----------------------GCGUGCUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGCAACGCGGUA ----.((((((((((..(((-((((((((((.((........)).)))----------------------))))))))))((...((((....)))).))))))).)))))......... ( -32.80) >DroYak_CAF1 162065 93 + 1 ----AGCAUGCAACCUAAUG-GGCGUGUCACUGGAAUCGCGCCGUGUG----------------------GCGUGCUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGCUGCGCGAAA ----(((((((((((..(((-(((((((((((((.......))).)))----------------------))))))))))((...((((....)))).))))))).))))))........ ( -33.20) >DroAna_CAF1 169285 96 + 1 --GUUGCAUGCAACCGACUGGGGCGUGUCAGUCGGUUCGCGCCAUGUG----------------------GCAUGUUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGGCACGGAGCA --(((.(..((((((((((((......)))))))))).))((((((.(----------------------((((....)))))(((((..((.....))..)))))))))))..).))). ( -34.80) >DroPer_CAF1 156598 119 + 1 UAUAUAUUUGCAACUAAAUG-GGCGUGUCAGUCGGUUUGCACCGUGUGUGUGUUCGUGUGUGGUGUGGCAGCGUGCUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGGCACGCGGCA ........(((....((((.-(((......))).))))((((((..(..(....)..)..)))))).)))((((((.((((.((...................)).)))))))))).... ( -36.01) >consensus ____AGCAUGCAACCUAAUG_GGCGUGUCACUCGAAUCGCGCCGUGUG______________________GCGUGCUCAUGUCAUAAUUCUAAAAUUUGCGGUUGUCAUGCUACGCGGAA .....((((((((((......((((((..........))))))...........................((((....))))..................))))).)))))......... (-21.34 = -21.28 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:45 2006