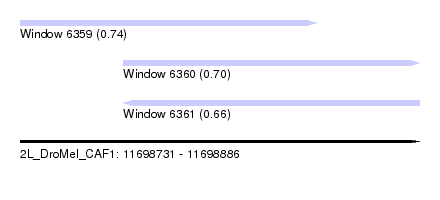
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,698,731 – 11,698,886 |
| Length | 155 |
| Max. P | 0.738168 |

| Location | 11,698,731 – 11,698,846 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 88.87 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11698731 115 + 22407834 UACAAUGCAUCACCUUAUGAACAUAUCGAGGCAGCUUUAAUGUCAACACCAAAUUUGCCUAAAGAGACUCCUUUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGG ............((...........((.((((((.(((..((....))..))).))))))...))...((((.((.....)).))))...))((((((((.....)))))))).. ( -28.30) >DroSec_CAF1 160204 112 + 1 UACAAUGCAACCCCU---GAACAUAUCGAGGCAGCUUUAAUGUCAACACCGAAUCUGCCUAAAGAGACUCCUGUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGG .............((---(......((.((((((.(((..((....))..))).))))))...))...(((((((.....)))))))..)))((((((((.....)))))))).. ( -36.90) >DroSim_CAF1 157771 115 + 1 UACAAUGCAACCCCUUAUGAACAUAUCGAGGCAGCUUUAAUGUCAACACCGAAUCUGCCUAAAGAGACUCCUGUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGG ............((...........((.((((((.(((..((....))..))).))))))...))...(((((((.....)))))))...))((((((((.....)))))))).. ( -36.60) >DroEre_CAF1 183107 115 + 1 UUUAAUGCGACCGCUCAUGAACCUAUUGAGACUGCUUUAAUGUCAACACCGAAUCUGCCUAAAGAGGCUCCUUUCAGCAGGACAGGAUACGGAUGAGCUCUACUUGGGCUCGUGG .((((.(((....((((.........))))..))).))))........(((.((((((((....))))((((......))))..)))).)))((((((((.....)))))))).. ( -32.40) >DroYak_CAF1 156966 115 + 1 AUCGAUGCAAAGCCUUAUGAACCUAUCGAGACAGCUUUAAUUUCAACACCGAAUCUGACUGAAGAGACUCCUCUCAGGCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGG .(((....(((((.((.(((.....))).))..)))))..........((...((((.((((.(((....))))))).))))..))...)))((((((((.....)))))))).. ( -31.50) >consensus UACAAUGCAACCCCUUAUGAACAUAUCGAGGCAGCUUUAAUGUCAACACCGAAUCUGCCUAAAGAGACUCCUGUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGG .........................(((((((((.(((..((....))..))).))))))........((((.((.....)).))))..)))((((((((.....)))))))).. (-23.80 = -23.80 + 0.00)

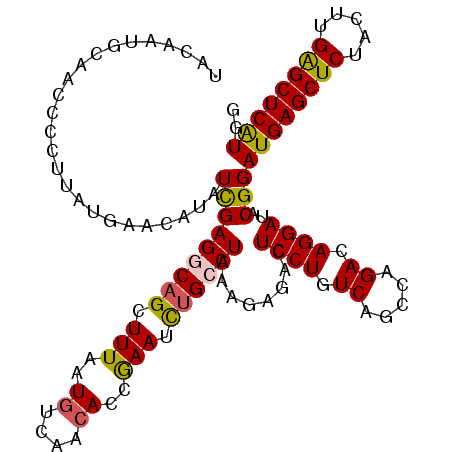
| Location | 11,698,771 – 11,698,886 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -23.01 |
| Energy contribution | -24.77 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11698771 115 + 22407834 UGUCAACACCAAAUUUGCCUAAAGAGACUCCUUUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGGUUAUGCAUCAGCUGCAUUUGCCGGUUCGGCCCAACCUGAG .............((((.((((((......)))).)).))))((((...(((((((((((.....)))))).((((.(((((.....)))))..)))))))))......)))).. ( -31.40) >DroSec_CAF1 160241 115 + 1 UGUCAACACCGAAUCUGCCUAAAGAGACUCCUGUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGGUUAUGCAUCAGCUGCAUUUGCCGGUUCGGCCCAACCUGGC .((((...(((..(((......)))...(((((((.....)))))))..)))((((((((.....))))))))(((((((((.....)))))..((((...))))..)))))))) ( -41.00) >DroSim_CAF1 157811 115 + 1 UGUCAACACCGAAUCUGCCUAAAGAGACUCCUGUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGGUUAUGCAUCAGCUGCAUUUGCCGGUUCGGCCCAACCUGGC .((((...(((..(((......)))...(((((((.....)))))))..)))((((((((.....))))))))(((((((((.....)))))..((((...))))..)))))))) ( -41.00) >DroEre_CAF1 183147 115 + 1 UGUCAACACCGAAUCUGCCUAAAGAGGCUCCUUUCAGCAGGACAGGAUACGGAUGAGCUCUACUUGGGCUCGUGGUUAUGCAUCAGCUGCAUUUGCCUGCUCGGCCCAGACUGGC .(((..(((((.((((((((....))))((((......))))..)))).))).)).........(((((.((.(((.(((((.....)))))..)))....)))))))))).... ( -38.00) >DroYak_CAF1 157006 115 + 1 UUUCAACACCGAAUCUGACUGAAGAGACUCCUCUCAGGCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGGUUAUGCAUCGGUUGCAUUUGCCUGUUCGGCCCAACCUGAC .............((((.((((.(((....))))))).))))((((...(((((((((((.....))))))..(((.(((((.....)))))..))).)))))......)))).. ( -39.50) >DroAna_CAF1 164459 87 + 1 ------------------------GAAAUCCUUU---ACAGACAGGAUGUGGAUGGCCU-UCGUUCAAUUCAUGGUUAUGCAUCGAUUGCAGUUGCCUGUUUGGCCAAAGUUGGC ------------------------..........---.((((((((((.((((((....-.))))))))))..(((..((((.....))))...)))))))))((((....)))) ( -23.80) >consensus UGUCAACACCGAAUCUGCCUAAAGAGACUCCUUUCAGCCAGACAGGAUACGGAUGAGCUCUACUUGAGCUCAUGGUUAUGCAUCAGCUGCAUUUGCCGGUUCGGCCCAACCUGGC ........((((((..............((((.((.....)).)))).....((((((((.....))))))))(((.(((((.....)))))..))).))))))........... (-23.01 = -24.77 + 1.75)

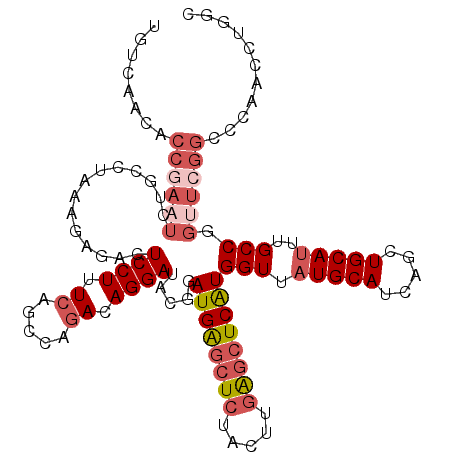
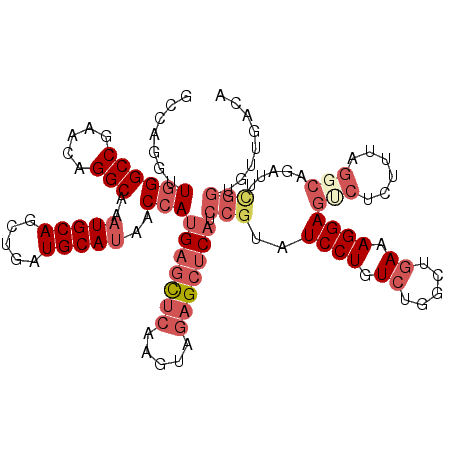
| Location | 11,698,771 – 11,698,886 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -22.43 |
| Energy contribution | -24.60 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11698771 115 - 22407834 CUCAGGUUGGGCCGAACCGGCAAAUGCAGCUGAUGCAUAACCAUGAGCUCAAGUAGAGCUCAUCCGUAUCCUGUCUGGCUGAAAGGAGUCUCUUUAGGCAAAUUUGGUGUUGACA .....((..(((((((.(((...(((((.....)))))....((((((((.....)))))))))))..((((.((.....)).))))((((....))))...)))))).)..)). ( -34.70) >DroSec_CAF1 160241 115 - 1 GCCAGGUUGGGCCGAACCGGCAAAUGCAGCUGAUGCAUAACCAUGAGCUCAAGUAGAGCUCAUCCGUAUCCUGUCUGGCUGACAGGAGUCUCUUUAGGCAGAUUCGGUGUUGACA .....((..(((((((.(((...(((((.....)))))....((((((((.....)))))))))))..(((((((.....)))))))((((....))))...)))))).)..)). ( -42.80) >DroSim_CAF1 157811 115 - 1 GCCAGGUUGGGCCGAACCGGCAAAUGCAGCUGAUGCAUAACCAUGAGCUCAAGUAGAGCUCAUCCGUAUCCUGUCUGGCUGACAGGAGUCUCUUUAGGCAGAUUCGGUGUUGACA .....((..(((((((.(((...(((((.....)))))....((((((((.....)))))))))))..(((((((.....)))))))((((....))))...)))))).)..)). ( -42.80) >DroEre_CAF1 183147 115 - 1 GCCAGUCUGGGCCGAGCAGGCAAAUGCAGCUGAUGCAUAACCACGAGCCCAAGUAGAGCUCAUCCGUAUCCUGUCCUGCUGAAAGGAGCCUCUUUAGGCAGAUUCGGUGUUGACA ....(((.(.((((((((((...(((((.....)))))......((((.(.....).))))........))))).(((((.(((((....))))).)))))..))))).).))). ( -38.00) >DroYak_CAF1 157006 115 - 1 GUCAGGUUGGGCCGAACAGGCAAAUGCAACCGAUGCAUAACCAUGAGCUCAAGUAGAGCUCAUCCGUAUCCUGUCUGCCUGAGAGGAGUCUCUUCAGUCAGAUUCGGUGUUGAAA ....(((((.(((.....)))...((((.....))))))))).(((((((.....)))))))((..((((..(((((.(((((((....))).)))).)))))..))))..)).. ( -41.20) >DroAna_CAF1 164459 87 - 1 GCCAACUUUGGCCAAACAGGCAACUGCAAUCGAUGCAUAACCAUGAAUUGAACGA-AGGCCAUCCACAUCCUGUCUGU---AAAGGAUUUC------------------------ ........(((((...(((....)))...(((((.(((....))).)))))....-.))))).....(((((......---..)))))...------------------------ ( -22.30) >consensus GCCAGGUUGGGCCGAACAGGCAAAUGCAGCUGAUGCAUAACCAUGAGCUCAAGUAGAGCUCAUCCGUAUCCUGUCUGGCUGAAAGGAGUCUCUUUAGGCAGAUUCGGUGUUGACA .......((((((.....)))..(((((.....)))))..)))(((((((.....))))))).(((..((((.((.....)).))))(((......))).....)))........ (-22.43 = -24.60 + 2.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:43 2006