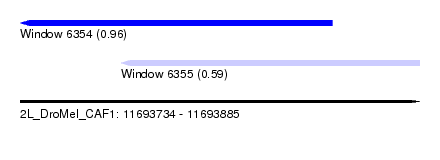
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,693,734 – 11,693,885 |
| Length | 151 |
| Max. P | 0.957642 |

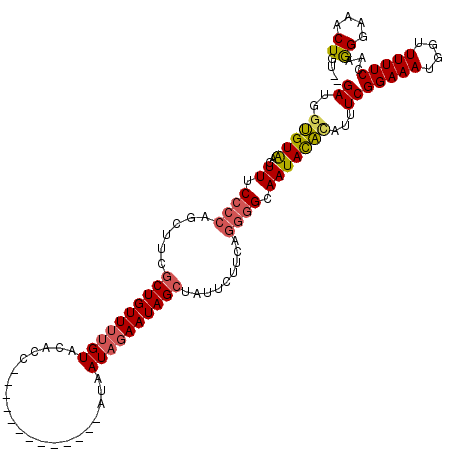
| Location | 11,693,734 – 11,693,852 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -28.62 |
| Consensus MFE | -16.82 |
| Energy contribution | -17.06 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11693734 118 - 22407834 GAUGGUGUGACUUUCCCCAGCUUCGCUGUUUUGUAUACUUAUAUAAUAGUAGAUAAUAGAAUAGCUAUUCUUCAGGGGCAAUACAUAUUUCGGAAAUAGUUUUUCAAAGGGAAACUGA-- ((..(((((..((.((((((..(.((((((((((.((((........))))....)))))))))).)..))...)))).)))))))...)).....((((((((.....)))))))).-- ( -27.20) >DroSec_CAF1 155197 105 - 1 GAUGGUGUAACUUUCCCCAGCAUCGCUGUUUUGUACACC-------------AUAAUAGAAUAGCUAUUCUUCAGGGGCAAUACACAUUUCGGAAAUGUUUUUUCCAAGGGAAACUGU-- ((..(((((..((.((((((.((.((((((((((.....-------------...)))))))))).)).))...)))).)))))))...))(((((.....)))))..((....))..-- ( -29.30) >DroSim_CAF1 152777 105 - 1 GAUGGUGUAACUUUCCCCAGCAUCGCUGUUUUGUACACC-------------AUAAUAGAAUAGCUAUUCUUCAGGGGCAAUACACAUUUCGGAAAUGGUUUUUCCAGGGGAAACUGU-- ((..(((((..((.((((((.((.((((((((((.....-------------...)))))))))).)).))...)))).)))))))...))(((((.....)))))..((....))..-- ( -29.40) >DroEre_CAF1 178255 93 - 1 GAUGGAGUAACUUUCUGCAGCUUCGCUGUUUUGUAGAGC-------------GUAAUAGAAUAG-------------GAAAUAUU-UAGUCGGAAAUGGUUUUUCGAGAGGAAACUGUUA (((.(((((.(((((((..((((..(......)..))))-------------....))))).))-------------....))))-).)))(((((....)))))((.((....)).)). ( -19.60) >DroYak_CAF1 151838 107 - 1 GAUGGAGUAACUUUCCCCUGCUUCGCUGUUUGGUAGACC-------------AUAAUAUAAUAGCGAUUCCUAAGGGGAAAUACUCUUUUCGGAAAUUGUUUUUCGAAAGGAAACUGUUA (((((.(((..((((((((...(((((((((((....))-------------)......))))))))......)))))))))))((((((((((((....))))))))))))..))))). ( -37.60) >consensus GAUGGUGUAACUUUCCCCAGCUUCGCUGUUUUGUACACC_____________AUAAUAGAAUAGCUAUUCUUCAGGGGCAAUACACAUUUCGGAAAUGGUUUUUCCAAGGGAAACUGU__ ((..(((((..((.((((......((((((((((.....................)))))))))).........)))).)))))))...))(((((....)))))...((....)).... (-16.82 = -17.06 + 0.24)


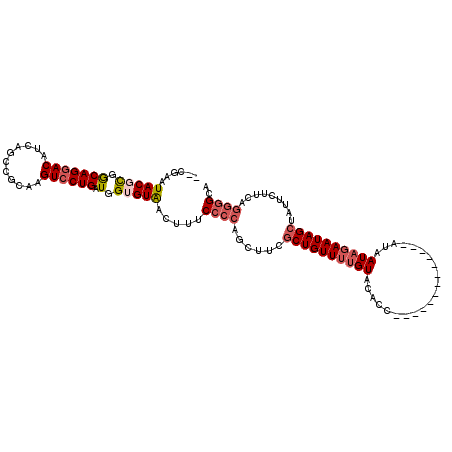
| Location | 11,693,772 – 11,693,885 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -18.62 |
| Energy contribution | -19.94 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11693772 113 - 22407834 --CGAAUACGCGGCAGGACAUCAGCCGCAAGUCCUGAUGGUGUGACUUUCCCCAGCUUCGCUGUUUUGUAUACUUAUAUAAUAGUAGAUAAUAGAAUAGCUAUUCUUCAGGGGCA --....(((((.(((((((...........)))))).).))))).....((((((..(.((((((((((.((((........))))....)))))))))).)..))...)))).. ( -31.60) >DroSec_CAF1 155235 100 - 1 --CGAAUACGCGGCAGGACAUCAGCCGCAAGUCCUGAUGGUGUAACUUUCCCCAGCAUCGCUGUUUUGUACACC-------------AUAAUAGAAUAGCUAUUCUUCAGGGGCA --.......(((((.........)))))..(((((((((.((..........)).))))((((((((((.....-------------...)))))))))).........))))). ( -31.20) >DroSim_CAF1 152815 100 - 1 --CGAAUACGUGGCAGGACAUCAGCCGCAAGUCCUGAUGGUGUAACUUUCCCCAGCAUCGCUGUUUUGUACACC-------------AUAAUAGAAUAGCUAUUCUUCAGGGGCA --.......(((((.........)))))..(((((((((.((..........)).))))((((((((((.....-------------...)))))))))).........))))). ( -29.30) >DroEre_CAF1 178294 89 - 1 UUGGGAUACGCGACAGGACAUCAGCUGUAAGUCCUGAUGGAGUAACUUUCUGCAGCUUCGCUGUUUUGUAGAGC-------------GUAAUAGAAUAG-------------GAA ......(((((.((((((((.(((((((((.(((....))).).......)))))))..).))))))))...))-------------))).........-------------... ( -25.41) >DroYak_CAF1 151878 100 - 1 --CGGAUACGCGGCAGGACAUCAGCUGUAAGUGCUGAUGGAGUAACUUUCCCCUGCUUCGCUGUUUGGUAGACC-------------AUAAUAUAAUAGCGAUUCCUAAGGGGAA --.(((.....((((((.(((((((.......)))))))(((.....))).))))))(((((((((((....))-------------)......)))))))).)))......... ( -33.20) >consensus __CGAAUACGCGGCAGGACAUCAGCCGCAAGUCCUGAUGGUGUAACUUUCCCCAGCUUCGCUGUUUUGUACACC_____________AUAAUAGAAUAGCUAUUCUUCAGGGGCA ......(((((.(((((((...........)))))).).))))).....((((......((((((((((.....................)))))))))).........)))).. (-18.62 = -19.94 + 1.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:37 2006