| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
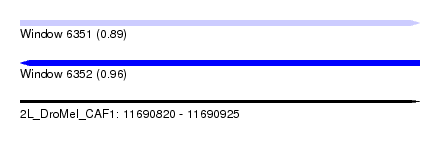
| Location | 11,690,820 – 11,690,925 |
| Length | 105 |
| Max. P | 0.963655 |

| Location | 11,690,820 – 11,690,925 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -23.93 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.02 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
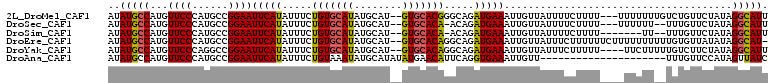
>2L_DroMel_CAF1 11690820 105 + 22407834 AUAUGCCAUGUUCCCAUGCCGGAAUUCAUAUUUCUGUGCAUAUGCAU--GUGCACGGGCAGAUGAAAUUGUUAUUUUCUUUU---UUUUUUUGUCUGUUCUAUAGGCAUU ..(((((..(((((......)))))..........(((((((....)--))))))(((((((..(((..(..(......)..---)..)))..)))))))....))))). ( -29.50) >DroSec_CAF1 152341 102 + 1 AUAUGCCAUGUUCCCAUGCCGGAAUUCAUAUUUCUGUGCAUAUGCAU--GUGCACA-ACAGAUGAAAUUGUUAUUUUCUUUU---UUUUUU--UUUGUUCUAUAGGCAUU ..(((((...((((......))))(((((.....((((((((....)--)))))))-....)))))................---......--...........))))). ( -22.30) >DroSim_CAF1 149919 98 + 1 AUAUGCCAUGUUCCCAUGCCGGAAUUCAUAUUUCUGUGCAUAUGCAU--GUGCACA-ACAGAUGAAAUUGUUAUUUUCUUUU-------UU--UUUGUUCUAUAGGCAUU ..(((((...((((......))))(((((.....((((((((....)--)))))))-....)))))................-------..--...........))))). ( -22.30) >DroEre_CAF1 175263 107 + 1 AUAUGCCAUGUUCCCAUGCCGGAAUUCAUAUUUCUGUGCAUAUGCAU--GUGCACAGGCAGAUGAAAUUGUUAUUUCUUUUUUCUUUUUUUUUUGUGUUAUAUAGGCAU- ..(((((((((..((((((((((((....)))))((((((((....)--)))))))))))...(((((....)))))................)).)..)))).)))))- ( -28.00) >DroYak_CAF1 148959 104 + 1 AUAUGCCAUGUUCCCAGGCCGGAAUUCAUAUUUCUGUGCAUAUGCAU--GUGCACAGGCAGAUGAAAUUGUUAUUUCUUUUU----UUCUUUUUGUCUUCUAUAGGCAUU ..(((((...((((......))))(((((...((((((((((....)--)))))))))...)))))................----..................))))). ( -26.80) >DroAna_CAF1 158322 89 + 1 AUAUGCCAUGUUCCCAUGCCGGAAUUCAUAUUUCUGUAAAUAUGCAUAUAUGAACAUUCAGGUGAAAUUGUU---------------------UUUGUUCCAUAGUUAUC ((((((.(((((....(((.(((((....))))).))))))))))))))..(((((..(((......)))..---------------------..))))).......... ( -14.70) >consensus AUAUGCCAUGUUCCCAUGCCGGAAUUCAUAUUUCUGUGCAUAUGCAU__GUGCACAGGCAGAUGAAAUUGUUAUUUUCUUUU____UUUUU__UUUGUUCUAUAGGCAUU ..(((((...((((......))))(((((.....(((((((........))))))).....)))))......................................))))). (-19.07 = -19.02 + -0.05)
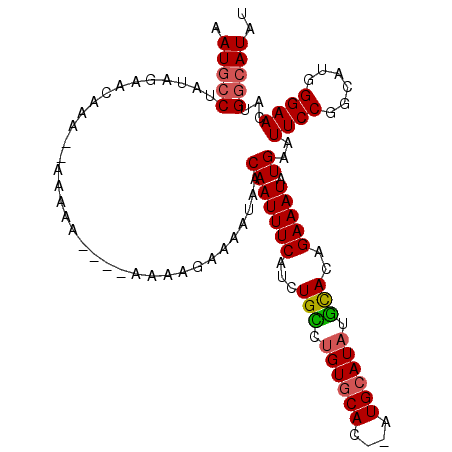
| Location | 11,690,820 – 11,690,925 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -21.18 |
| Consensus MFE | -18.33 |
| Energy contribution | -18.42 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
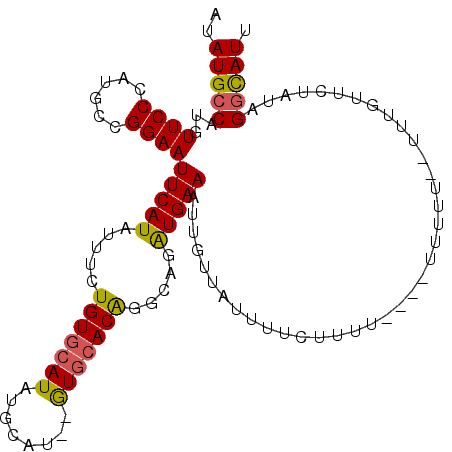
>2L_DroMel_CAF1 11690820 105 - 22407834 AAUGCCUAUAGAACAGACAAAAAAA---AAAAGAAAAUAACAAUUUCAUCUGCCCGUGCAC--AUGCAUAUGCACAGAAAUAUGAAUUCCGGCAUGGGAACAUGGCAUAU .(((((...................---....((((.......)))).....(((((((..--...(((((........))))).......))))))).....))))).. ( -23.60) >DroSec_CAF1 152341 102 - 1 AAUGCCUAUAGAACAAA--AAAAAA---AAAAGAAAAUAACAAUUUCAUCUGU-UGUGCAC--AUGCAUAUGCACAGAAAUAUGAAUUCCGGCAUGGGAACAUGGCAUAU .(((((...........--......---................(((((...(-((((((.--.......)))))))....)))))((((......))))...))))).. ( -20.90) >DroSim_CAF1 149919 98 - 1 AAUGCCUAUAGAACAAA--AA-------AAAAGAAAAUAACAAUUUCAUCUGU-UGUGCAC--AUGCAUAUGCACAGAAAUAUGAAUUCCGGCAUGGGAACAUGGCAUAU .(((((...........--..-------................(((((...(-((((((.--.......)))))))....)))))((((......))))...))))).. ( -20.90) >DroEre_CAF1 175263 107 - 1 -AUGCCUAUAUAACACAAAAAAAAAAGAAAAAAGAAAUAACAAUUUCAUCUGCCUGUGCAC--AUGCAUAUGCACAGAAAUAUGAAUUCCGGCAUGGGAACAUGGCAUAU -((((((((((......................(((((....)))))......(((((((.--.......)))))))..)))))..((((......))))...))))).. ( -22.60) >DroYak_CAF1 148959 104 - 1 AAUGCCUAUAGAAGACAAAAAGAA----AAAAAGAAAUAACAAUUUCAUCUGCCUGUGCAC--AUGCAUAUGCACAGAAAUAUGAAUUCCGGCCUGGGAACAUGGCAUAU .(((((..................----................(((((.(..(((((((.--.......)))))))..).)))))((((......))))...))))).. ( -22.90) >DroAna_CAF1 158322 89 - 1 GAUAACUAUGGAACAAA---------------------AACAAUUUCACCUGAAUGUUCAUAUAUGCAUAUUUACAGAAAUAUGAAUUCCGGCAUGGGAACAUGGCAUAU .........((((....---------------------((((..(((....)))))))........(((((((....)))))))..))))(.((((....)))).).... ( -16.20) >consensus AAUGCCUAUAGAACAAA__AAAAA____AAAAGAAAAUAACAAUUUCAUCUGCCUGUGCAC__AUGCAUAUGCACAGAAAUAUGAAUUCCGGCAUGGGAACAUGGCAUAU .(((((..................................(((((((...(((.((((((....)))))).)))..))))).))..((((......))))...))))).. (-18.33 = -18.42 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:34 2006