
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,669,170 – 11,669,318 |
| Length | 148 |
| Max. P | 0.963411 |

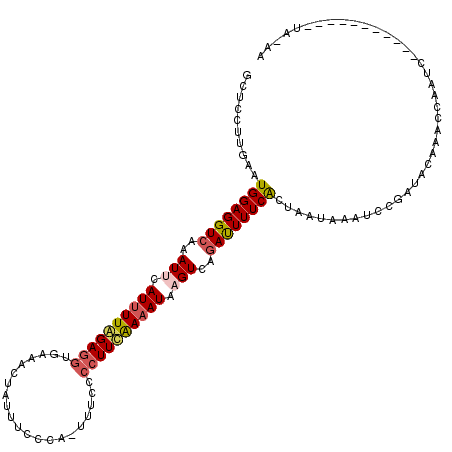
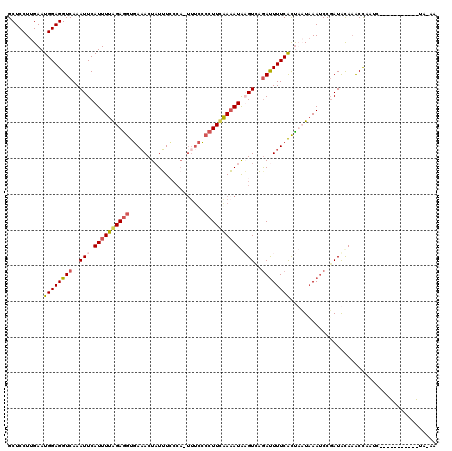
| Location | 11,669,170 – 11,669,278 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.06 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -11.61 |
| Energy contribution | -12.55 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11669170 108 - 22407834 GCUCCUUGAAUGGAGGUCAAAUUCAUUUUAGAAAUGAAACUAUUUCCCA-UUUCCCCUUUGAAAUAUGUCAGAUUUUCAUUACUAAACCCGAUACAAACCAAUC-----------UAAAU ..(((......)))(((.....((..(((((.(((((((..((((..((-((((......)))..)))..))))))))))).)))))...)).....)))....-----------..... ( -15.10) >DroSec_CAF1 130409 119 - 1 GCUCCUUGAAUGGAGGUCAAAUUCAUUUUGGAGGUGAAACUAUUUCCCA-UUUCCCCUUCAAAAUAAGUCAAACUUUCGCUUAUAAAUCCUAUACAAACCUAUCACAGAGCUAUUUAAAA ((((..(((..((((((...(((.((((((((((.((((..........-)))).)))))))))).)))...))))))...((((.....))))........)))..))))......... ( -24.50) >DroSim_CAF1 128303 93 - 1 GCUCCUUGAAUGGAGGUCAAAUUCAUUUUGGAGGUGAAACUAUUU--------CCCCUUCAAAAUAAGUCAGAUUUUCACUUAUAAAUCCGAUAUAUACUU------------------- .((((......)))).........((((((((((.(((.....))--------).))))))))))..(((.(((((........))))).)))........------------------- ( -22.40) >DroYak_CAF1 126216 119 - 1 GCUCCUUGAAUGGAGGUCAAAUUCAUUUUAGAGGUUAUACUUUAUACCAUUUUUGCCUUAAAUAUGUGUUUGAUUUUCACCACUCAAUCCGAUGCAAGCCAAUGACAAAACUAUUUA-AA ((((.((((.((((((((((((.(((.((.(((((...................))))).)).))).))))))))))))....))))...)).))......................-.. ( -20.81) >consensus GCUCCUUGAAUGGAGGUCAAAUUCAUUUUAGAGGUGAAACUAUUUCCCA_UUUCCCCUUCAAAAUAAGUCAGAUUUUCACUAAUAAAUCCGAUACAAACCAAUC___________UA_AA ..........((((((((..(((.((((((((((.....................)))))))))).)))..))))))))......................................... (-11.61 = -12.55 + 0.94)

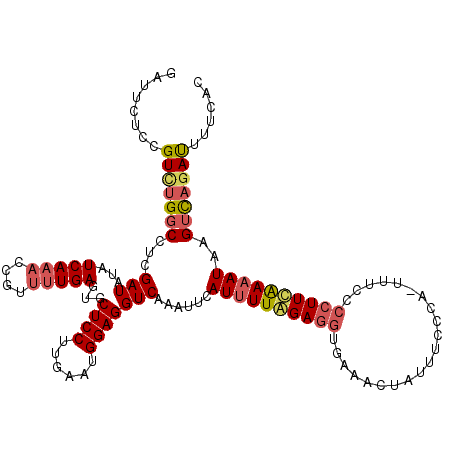
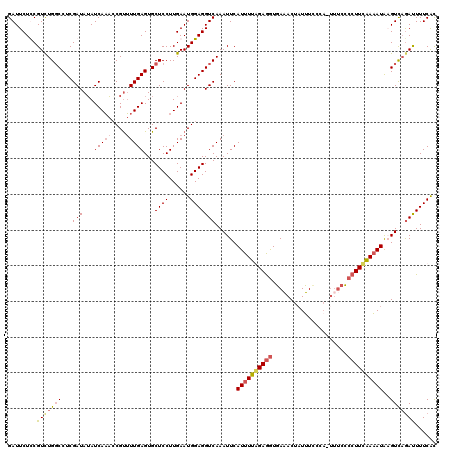
| Location | 11,669,199 – 11,669,318 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.54 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -17.39 |
| Energy contribution | -18.70 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11669199 119 - 22407834 GAUUCUUCGUCUGGCCUCGAUAUAUCAAACCGUUUUGAGUGCUCCUUGAAUGGAGGUCAAAUUCAUUUUAGAAAUGAAACUAUUUCCCA-UUUCCCCUUUGAAAUAUGUCAGAUUUUCAU ........(((((((...(((...(((((....)))))...((((......)))))))...((((.....((((((...........))-)))).....))))....)))))))...... ( -23.10) >DroSec_CAF1 130449 119 - 1 GAUUCUCCGUCUGUCCUCGAUAUAUCAAAACGUUUUGAGUGCUCCUUGAAUGGAGGUCAAAUUCAUUUUGGAGGUGAAACUAUUUCCCA-UUUCCCCUUCAAAAUAAGUCAAACUUUCGC (((.(((((((.(..(((((.((........)).)))))..).....).)))))))))......((((((((((.((((..........-)))).))))))))))............... ( -25.40) >DroSim_CAF1 128324 112 - 1 GAUUCUCCGUUUGGCCUCGAUAUAUCAAACCGUUUUGAGUGCUCCUUGAAUGGAGGUCAAAUUCAUUUUGGAGGUGAAACUAUUU--------CCCCUUCAAAAUAAGUCAGAUUUUCAC ....((((((((((((((((.((........)).))))).)))....))))))))(((..(((.((((((((((.(((.....))--------).)))))))))).)))..)))...... ( -30.50) >DroYak_CAF1 126255 120 - 1 GAUUCUUCGUCUGGCCUCGAUAGAUCAUCCCGCAUUGAGUGCUCCUUGAAUGGAGGUCAAAUUCAUUUUAGAGGUUAUACUUUAUACCAUUUUUGCCUUAAAUAUGUGUUUGAUUUUCAC .....((((...(((((((((.(.......)..)))))).)))...))))((((((((((((.(((.((.(((((...................))))).)).))).)))))))))))). ( -27.71) >consensus GAUUCUCCGUCUGGCCUCGAUAUAUCAAACCGUUUUGAGUGCUCCUUGAAUGGAGGUCAAAUUCAUUUUAGAGGUGAAACUAUUUCCCA_UUUCCCCUUCAAAAUAAGUCAGAUUUUCAC ........(((((((...(((...(((((....)))))...((((......)))))))......((((((((((.....................))))))))))..)))))))...... (-17.39 = -18.70 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:20 2006