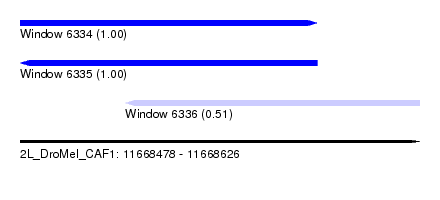
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,668,478 – 11,668,626 |
| Length | 148 |
| Max. P | 0.999949 |

| Location | 11,668,478 – 11,668,588 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 97.45 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -23.94 |
| Energy contribution | -24.14 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
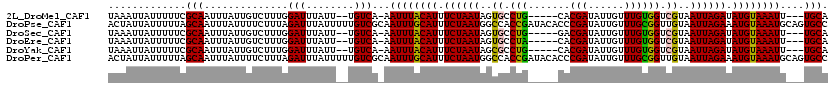
>2L_DroMel_CAF1 11668478 110 + 22407834 CAUGAAGUAUUUAUCUCUCGACCAUCGGGCCAUACAAAUUGCAAAUUUACAUAUCUAAUUACGACCACAAACAAUAUCGUGCAGGCACUAUUAGAAAUGUAAAUUUGACA ......((((......((((.....))))..))))....(((((((((((((.((((((...(.((....((......))...)))...)))))).))))))))))).)) ( -21.90) >DroSec_CAF1 129724 110 + 1 CAUGAAGUAUUUAUCUCUCGGCCAUCGGGCCGUACAAAUUGCAAAUUUACAUAUCUAAUUACGACCACAAACAAUAUCGUCCAGGCACUAUUAGAAAUGUAAAUUUGACA ......((..........(((((....))))).........(((((((((((.((((((...(.((....((......))...)))...)))))).))))))))))))). ( -25.70) >DroSim_CAF1 127624 110 + 1 CAUGAAGUAUUUAUCUCUCGGCCAUCGGGCCAUACAAAUUGCAAAUUUACAUAUCUAAUUACGACCACAAACAAUAUCGUGCAGGCACUAUUAGAAAUGUAAAUUUGACA ......((...........((((....))))..........(((((((((((.((((((...(.((....((......))...)))...)))))).))))))))))))). ( -25.70) >DroEre_CAF1 151214 110 + 1 CAUGAAGUAUUUAUCUAUCGGCCAUCGGGCCAUGCAAAUUGCAAAUUUACAUAUCUAAUUACGACCACAAACAAUAUCGUGUAGGCACUAUUAGAAAUGUAAAUUUGACA ......((...........((((....))))..........(((((((((((.((((((...(.(((((..........))).)))...)))))).))))))))))))). ( -26.10) >DroYak_CAF1 125533 110 + 1 CAUGAAGUAUUUAUCUCUCGGCCAUCGGGCCAUACAAAUUGCAAAUUUACAUAUCUAAUUACGACCACAAACAAUAUCGUGCAGGCGCUAUUAGAAAUGUAAAUUUGACA ......((...........((((....))))..........(((((((((((.((((((..((.((....((......))...))))..)))))).))))))))))))). ( -29.10) >consensus CAUGAAGUAUUUAUCUCUCGGCCAUCGGGCCAUACAAAUUGCAAAUUUACAUAUCUAAUUACGACCACAAACAAUAUCGUGCAGGCACUAUUAGAAAUGUAAAUUUGACA ......((...........((((....))))..........(((((((((((.((((((...(.((....((......))...)))...)))))).))))))))))))). (-23.94 = -24.14 + 0.20)

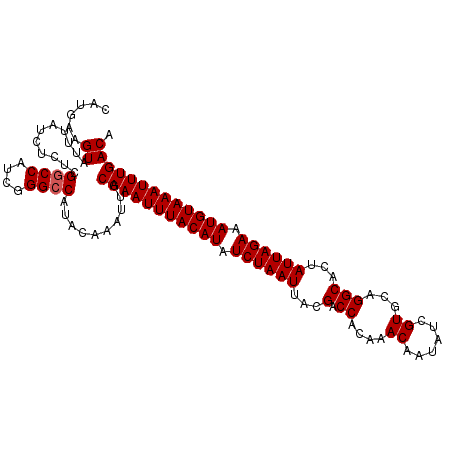

| Location | 11,668,478 – 11,668,588 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 97.45 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.22 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.78 |
| SVM RNA-class probability | 0.999949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11668478 110 - 22407834 UGUCAAAUUUACAUUUCUAAUAGUGCCUGCACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUUUGCAAUUUGUAUGGCCCGAUGGUCGAGAGAUAAAUACUUCAUG ((.(((((((((((.((((((..(((...(((((......)))))...))))))))).)))))))))))))((((((.(((((....)))))....))))))........ ( -28.30) >DroSec_CAF1 129724 110 - 1 UGUCAAAUUUACAUUUCUAAUAGUGCCUGGACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUUUGCAAUUUGUACGGCCCGAUGGCCGAGAGAUAAAUACUUCAUG ((.(((((((((((.((((((..(((((..(((....)))..).)).))..)))))).)))))))))))))((((((.(((((....)))))....))))))........ ( -33.50) >DroSim_CAF1 127624 110 - 1 UGUCAAAUUUACAUUUCUAAUAGUGCCUGCACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUUUGCAAUUUGUAUGGCCCGAUGGCCGAGAGAUAAAUACUUCAUG ((.(((((((((((.((((((..(((...(((((......)))))...))))))))).)))))))))))))((((((.(((((....)))))....))))))........ ( -31.00) >DroEre_CAF1 151214 110 - 1 UGUCAAAUUUACAUUUCUAAUAGUGCCUACACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUUUGCAAUUUGCAUGGCCCGAUGGCCGAUAGAUAAAUACUUCAUG ((.(((((((((((.((((((..(((...(((((......)))))...))))))))).)))))))))))))(((((..(((((....))))).)))))............ ( -30.80) >DroYak_CAF1 125533 110 - 1 UGUCAAAUUUACAUUUCUAAUAGCGCCUGCACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUUUGCAAUUUGUAUGGCCCGAUGGCCGAGAGAUAAAUACUUCAUG ((.(((((((((((.((((((.(((((...((((......)))))).))).)))))).)))))))))))))((((((.(((((....)))))....))))))........ ( -33.60) >consensus UGUCAAAUUUACAUUUCUAAUAGUGCCUGCACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUUUGCAAUUUGUAUGGCCCGAUGGCCGAGAGAUAAAUACUUCAUG ((.(((((((((((.((((((..((((...((((......)))))).))..)))))).)))))))))))))((((((.(((((....)))))....))))))........ (-30.50 = -30.22 + -0.28)

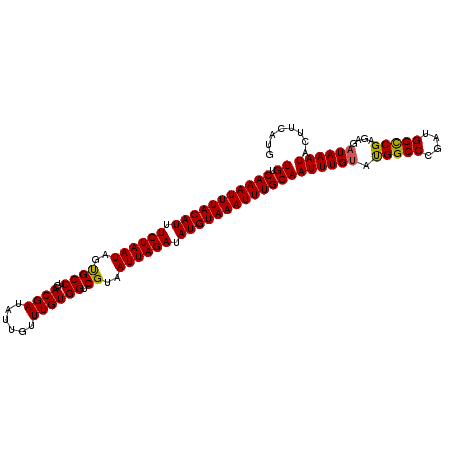
| Location | 11,668,517 – 11,668,626 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.52 |
| Mean single sequence MFE | -26.85 |
| Consensus MFE | -17.06 |
| Energy contribution | -16.34 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11668517 109 - 22407834 UAAAUUAUUUUUCGCAAUUUAUUGUCUUUGGAUUUAUU--UGUCA-AAUUUACAUUUCUAAUAGUGCCUG-----CACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUU---UGCA ((((((.......((((....)))).....))))))..--((.((-(((((((((.((((((..(((...-----(((((......)))))...))))))))).)))))))))---)))) ( -23.40) >DroPse_CAF1 119304 120 - 1 ACUAUUAUUUUUAGCAAUUUAUUUUCUUUAGAUUUAUUUUUGUCGCAAUUUGCAUUUCUAAUGGCCACCGAUACACCCGAUAUUGUUUGCGGUUGUAAUUAGAAAUGUAAAUGCAGUGCC .............(((..............(((........)))(((.((((((((((((((.((.((((..(((........)))...)))).)).)))))))))))))))))..))). ( -31.50) >DroSec_CAF1 129763 109 - 1 UAAAUUAUUUUUCGCAAUUUAUUGUCUUUGGAUUUAUU--UGUCA-AAUUUACAUUUCUAAUAGUGCCUG-----GACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUU---UGCA ((((((.......((((....)))).....))))))..--((.((-(((((((((.((((((..(((((.-----.(((....)))..).)).))..)))))).)))))))))---)))) ( -24.20) >DroEre_CAF1 151253 109 - 1 UAAAUUAUUUUUCGCAAUUUAUUGUCUUUGGAUUUAUU--UGUCA-AAUUUACAUUUCUAAUAGUGCCUA-----CACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUU---UGCA ((((((.......((((....)))).....))))))..--((.((-(((((((((.((((((..(((...-----(((((......)))))...))))))))).)))))))))---)))) ( -24.50) >DroYak_CAF1 125572 109 - 1 UAAAUUAUUUUUCGCAAUUUAUUGUCUUUGGAUUUAUU--UGUCA-AAUUUACAUUUCUAAUAGCGCCUG-----CACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUU---UGCA ((((((.......((((....)))).....))))))..--((.((-(((((((((.((((((.(((((..-----.((((......)))))).))).)))))).)))))))))---)))) ( -26.00) >DroPer_CAF1 120857 120 - 1 ACUAUUAUUUUUAGCAAUUUAUUUUCUUUAGAUUUAUUUUUGUCGCAAUUUGCAUUUCUAAUGGCCACCGAUACACCCGAUAUUGUUUGCGGUUGUAAUUAGAAAUGUAAAUGCAGUGCC .............(((..............(((........)))(((.((((((((((((((.((.((((..(((........)))...)))).)).)))))))))))))))))..))). ( -31.50) >consensus UAAAUUAUUUUUCGCAAUUUAUUGUCUUUGGAUUUAUU__UGUCA_AAUUUACAUUUCUAAUAGCGCCUG_____CACGAUAUUGUUUGUGGUCGUAAUUAGAUAUGUAAAUU___UGCA .............(((..............(((........)))...((((((((.((((((..((.(((.......(((......)))))).))..)))))).))))))))....))). (-17.06 = -16.34 + -0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:18 2006