
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
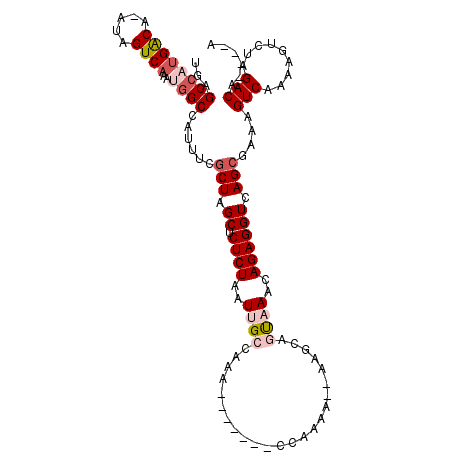
| Location | 11,662,957 – 11,663,057 |
| Length | 100 |
| Max. P | 0.696191 |

| Location | 11,662,957 – 11,663,057 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -21.25 |
| Consensus MFE | -14.25 |
| Energy contribution | -15.48 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620248 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
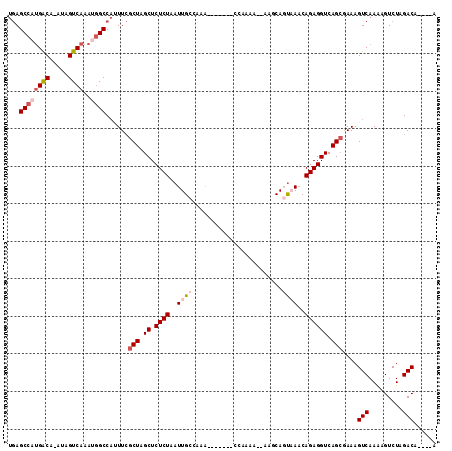
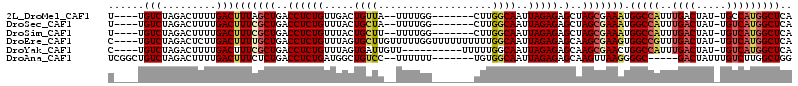
>2L_DroMel_CAF1 11662957 100 + 22407834 UGAGCCAUGGCA-AUAGUCAAAUGGCCAUUUCGCUAGCUCUCUAAUUGCCAAG-------CCAAAA--UAACAGUCAACAGAGGUCAGCUAAAGUCAAAAGUCUAGACA----A ((.((..(((((-((((.....((((......)))).....)).))))))).)-------)))...--....(((..((....))..)))...(((.........))).----. ( -18.30) >DroSec_CAF1 124287 100 + 1 UGAGCCAUGACA-AUAGUCAAAUGGCCAUUUCGCUAGCUCUCUAAUUGCCAAG-------CCAAAA--UAGCAGUAAACAGAGGUCAGCGAAAGUCAAAAGUCUAGACA----A ((.((((((((.-...))))..))))))(((((((..(.((((..((((...(-------(.....--..)).))))..)))))..)))))))(((.........))).----. ( -28.10) >DroSim_CAF1 122100 100 + 1 UGAGCCAUGACA-AUAGUCAAAUGGCCAUUUCGCUAGCUCUCUAAUUGCCAAG-------CCAAAA--AAGCAGUAAACAGAGGUCAGCGAAAGUCAAAAGUCUAGACA----A ((.((((((((.-...))))..))))))(((((((..(.((((..((((...(-------(.....--..)).))))..)))))..)))))))(((.........))).----. ( -28.10) >DroEre_CAF1 145775 109 + 1 UGAGCCAUGACA-AUAGUCAAACGGCCACUUCGCUUGCUCUCUAAUUGCCAAAAAAAAAACCAAAAACAAGCACUAAACAGAGGUCAGCAAAAGUCAAGAGUCUAGACA----G ((.(((.((((.-...))))...)))))....(((.((.((((...(((.....................)))......)))))).)))....(((.((...)).))).----. ( -17.60) >DroYak_CAF1 120034 99 + 1 UGAGCCAUGACA-AUAGUCAAAUGGCCAGUUCGCUUGCUCUCUAAUUGCCAAAAA----------AACAAUCACUAAACAGAGGUCAGCGAAAGUCAAAAGUCUAGACA----G ((.((((((((.-...))))..)))))).((((((.((.((((............----------..............)))))).)))))).(((.........))).----. ( -22.47) >DroAna_CAF1 130216 100 + 1 CCAGCCAAGACAAAUAGUC-----GCCCCUUAACUUGCUCUCUAAUUGCCACA-------AAAAAA--GGACAGCCAUCAGAGGUCAGAGAAAGUCAAAAGUCUAGACAGCCGA ................(((-----....(((.((((..(((((...((((...-------......--)).))(((......))).)))))))))...)))....)))...... ( -12.90) >consensus UGAGCCAUGACA_AUAGUCAAAUGGCCAUUUCGCUAGCUCUCUAAUUGCCAAA_______CCAAAA__AAGCAGUAAACAGAGGUCAGCGAAAGUCAAAAGUCUAGACA____A ...((((((((.....))))..))))......(((.((.((((..((((........................))))..)))))).)))....(((.........)))...... (-14.25 = -15.48 + 1.22)

| Location | 11,662,957 – 11,663,057 |
|---|---|
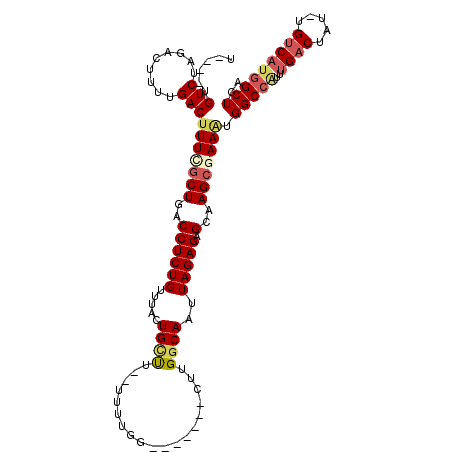
| Length | 100 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -27.05 |
| Consensus MFE | -19.94 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11662957 100 - 22407834 U----UGUCUAGACUUUUGACUUUAGCUGACCUCUGUUGACUGUUA--UUUUGG-------CUUGGCAAUUAGAGAGCUAGCGAAAUGGCCAUUUGACUAU-UGCCAUGGCUCA (----((((.((.((..((((.(((((........)))))..))))--....))-------)).))))).....((((((....(((((.(....).))))-)....)))))). ( -23.30) >DroSec_CAF1 124287 100 - 1 U----UGUCUAGACUUUUGACUUUCGCUGACCUCUGUUUACUGCUA--UUUUGG-------CUUGGCAAUUAGAGAGCUAGCGAAAUGGCCAUUUGACUAU-UGUCAUGGCUCA .----.(((.........)))((((((((.((((((.....(((((--......-------..)))))..))))).).)))))))).(((((..((((...-.))))))))).. ( -30.40) >DroSim_CAF1 122100 100 - 1 U----UGUCUAGACUUUUGACUUUCGCUGACCUCUGUUUACUGCUU--UUUUGG-------CUUGGCAAUUAGAGAGCUAGCGAAAUGGCCAUUUGACUAU-UGUCAUGGCUCA .----.(((.........)))((((((((.((((((....(.(((.--....))-------)..).....))))).).)))))))).(((((..((((...-.))))))))).. ( -28.40) >DroEre_CAF1 145775 109 - 1 C----UGUCUAGACUCUUGACUUUUGCUGACCUCUGUUUAGUGCUUGUUUUUGGUUUUUUUUUUGGCAAUUAGAGAGCAAGCGAAGUGGCCGUUUGACUAU-UGUCAUGGCUCA .----..............(((((.(((((.......)))))((((((((((.(((........))).....)))))))))))))))(((((..((((...-.))))))))).. ( -25.50) >DroYak_CAF1 120034 99 - 1 C----UGUCUAGACUUUUGACUUUCGCUGACCUCUGUUUAGUGAUUGUU----------UUUUUGGCAAUUAGAGAGCAAGCGAACUGGCCAUUUGACUAU-UGUCAUGGCUCA .----.(((.........))).((((((...((((.(.....(((((((----------.....)))))))).))))..))))))..(((((..((((...-.))))))))).. ( -26.50) >DroAna_CAF1 130216 100 - 1 UCGGCUGUCUAGACUUUUGACUUUCUCUGACCUCUGAUGGCUGUCC--UUUUUU-------UGUGGCAAUUAGAGAGCAAGUUAAGGGGC-----GACUAUUUGUCUUGGCUGG .(((((.......(((((((((((((((((((......)).(((((--......-------.).)))).)))))))..))))))))))..-----(((.....)))..))))). ( -28.20) >consensus U____UGUCUAGACUUUUGACUUUCGCUGACCUCUGUUUACUGCUU__UUUUGG_______CUUGGCAAUUAGAGAGCAAGCGAAAUGGCCAUUUGACUAU_UGUCAUGGCUCA ......(((.........)))(((((((..((((((.....((((...................))))..))))).)..))))))).(((((..((((.....))))))))).. (-19.94 = -20.78 + 0.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:12 2006