
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,660,468 – 11,660,565 |
| Length | 97 |
| Max. P | 0.658289 |

| Location | 11,660,468 – 11,660,565 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -17.62 |
| Consensus MFE | -10.90 |
| Energy contribution | -12.35 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
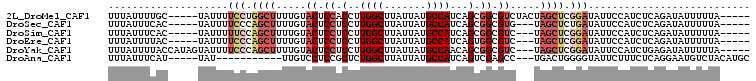
>2L_DroMel_CAF1 11660468 97 + 22407834 UUUAUUUUGC-----UAUUUUCCUGGCUUUUGUACUCCACCUUGGCUUAUUAUGCCAUCAGCGGCGUCUACUAGCUCGGAUAUUCCAUCUCAGAUAUUUUUA----- ..........-----.....(((.((((..((.((.((.(..((((.......))))...).)).)).))..)))).)))......................----- ( -18.70) >DroSec_CAF1 121783 94 + 1 UUUAUUUCAC-----UAUUUUCCCAGCUUUUGUACUCCUCCUUGGCUUAUUAUGCCAUCAGCGGCGUG---UAGCUCUGAUAUUCCAUCUCAGAUAUUUUUA----- ..........-----..........(((....(((.((.(..((((.......))))...).)).)))---.)))(((((.........)))))........----- ( -15.10) >DroSim_CAF1 119609 94 + 1 UUUAUUUCAC-----UAUUUUUCCAGCUUUUGUACUCCUCCUUGGCUUAUUAUGCCAUCAGCGGCGUC---UAGCUCGGAUAUUCCAUCUCAGAUAUUUUUA----- ..........-----......(((((((...(.((.((.(..((((.......))))...).)).)))---.)))).)))......................----- ( -16.60) >DroEre_CAF1 143306 94 + 1 UUUAUUUUAC-----UAUUUUCCCAGCUUUUGUACUCCUCCUUGGCUUAUUAUGCCAUCAGUGGCGUC---UAGCUCGGAUAUUCCAUCUCAGAUAUUUUUA----- ..........-----.....(((.((((...(.((.((.(..((((.......))))...).)).)))---.)))).)))......................----- ( -17.50) >DroYak_CAF1 117531 99 + 1 UUUAUUUUACCAUAGUAUUUUCCCAGCUUUUGUACUCCUCCUUGGCUUAUUAUGCCAACAGCGGCGUC---UAGCUCGGAUAUUCCAUCUGAGAUAUUUUUA----- ..(((((((...........(((.((((...(.((.((.(.(((((.......)))))..).)).)))---.)))).))).........)))))))......----- ( -19.05) >DroAna_CAF1 127619 88 + 1 UUUAUUUCAU-----UAU-----------UUGUCCUCCGCUCUGGCUUAUUAUGCCAUCAGUCGAGCC---UGACUGGGGUAUUCUUUCUCAGGAAUGUCUACAUGC .......(((-----(..-----------.....(((.(((.((((.......))))..))).)))((---(((..(((.....)))..)))))))))......... ( -18.80) >consensus UUUAUUUCAC_____UAUUUUCCCAGCUUUUGUACUCCUCCUUGGCUUAUUAUGCCAUCAGCGGCGUC___UAGCUCGGAUAUUCCAUCUCAGAUAUUUUUA_____ ....................(((.((((.....((.((.(..((((.......))))...).)).)).....)))).)))........................... (-10.90 = -12.35 + 1.45)

| Location | 11,660,468 – 11,660,565 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -19.24 |
| Consensus MFE | -12.17 |
| Energy contribution | -14.03 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11660468 97 - 22407834 -----UAAAAAUAUCUGAGAUGGAAUAUCCGAGCUAGUAGACGCCGCUGAUGGCAUAAUAAGCCAAGGUGGAGUACAAAAGCCAGGAAAAUA-----GCAAAAUAAA -----........(((.....)))...(((..(((.....((.(((((..((((.......)))).))))).)).....)))..))).....-----.......... ( -22.50) >DroSec_CAF1 121783 94 - 1 -----UAAAAAUAUCUGAGAUGGAAUAUCAGAGCUA---CACGCCGCUGAUGGCAUAAUAAGCCAAGGAGGAGUACAAAAGCUGGGAAAAUA-----GUGAAAUAAA -----........(((((.........)))))....---(((.((.((..((((.......)))).)).))(((......))).........-----)))....... ( -16.50) >DroSim_CAF1 119609 94 - 1 -----UAAAAAUAUCUGAGAUGGAAUAUCCGAGCUA---GACGCCGCUGAUGGCAUAAUAAGCCAAGGAGGAGUACAAAAGCUGGAAAAAUA-----GUGAAAUAAA -----........(((.....)))...(((.((((.---.((.((.((..((((.......)))).)).)).)).....)))))))......-----.......... ( -18.10) >DroEre_CAF1 143306 94 - 1 -----UAAAAAUAUCUGAGAUGGAAUAUCCGAGCUA---GACGCCACUGAUGGCAUAAUAAGCCAAGGAGGAGUACAAAAGCUGGGAAAAUA-----GUAAAAUAAA -----........(((.....)))...(((.((((.---.((.((.((..((((.......)))).)).)).)).....)))).))).....-----.......... ( -20.30) >DroYak_CAF1 117531 99 - 1 -----UAAAAAUAUCUCAGAUGGAAUAUCCGAGCUA---GACGCCGCUGUUGGCAUAAUAAGCCAAGGAGGAGUACAAAAGCUGGGAAAAUACUAUGGUAAAAUAAA -----......((((..((.((.....(((.((((.---.((.((.((.(((((.......))))))).)).)).....)))).)))...))))..))))....... ( -21.70) >DroAna_CAF1 127619 88 - 1 GCAUGUAGACAUUCCUGAGAAAGAAUACCCCAGUCA---GGCUCGACUGAUGGCAUAAUAAGCCAGAGCGGAGGACAA-----------AUA-----AUGAAAUAAA .(((.......((((...............(((((.---.....))))).((((.......))))....)))).....-----------...-----)))....... ( -16.36) >consensus _____UAAAAAUAUCUGAGAUGGAAUAUCCGAGCUA___GACGCCGCUGAUGGCAUAAUAAGCCAAGGAGGAGUACAAAAGCUGGGAAAAUA_____GUAAAAUAAA ...........................(((.((((.....((.((.((..((((.......)))).)).)).)).....)))).))).................... (-12.17 = -14.03 + 1.86)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:09 2006