| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,648,042 – 11,648,145 |
| Length | 103 |
| Max. P | 0.905038 |

| Location | 11,648,042 – 11,648,145 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.43 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -20.57 |
| Energy contribution | -21.18 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
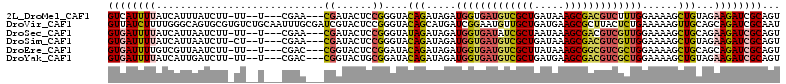
>2L_DroMel_CAF1 11648042 103 + 22407834 GUCAUUUUAUCAUUUAUCUU-UU--U---CGAA---CGAUACUCCGGGUACAGAUAGAUGGUGAUGUCGCUGAUAAAGCGACGUCUUUGGAAAAGCUGUAGAAGAUCGCAGU ........(((((((((((.-.(--(---((..---........))))...)))))))))))(((((((((.....))))))))).........(((((........))))) ( -30.10) >DroVir_CAF1 9582 112 + 1 GUUAUCUUUUGGGCAGUGCGUGUCUGCAAUUUGCGAUCGUACUCCGGGUACAGCAUGAUCGGAAUGUUGCUGAUGAAGCGCUUACUCUGAAAAAGUUGCAGCAGAUCGCAAU ............((((.......))))...((((((((((((.....)))).(((...(((((.((.((((.....))))..)).)))))......)))....)))))))). ( -31.40) >DroSec_CAF1 101450 103 + 1 GUGAUUUUAUCAUUAAUCUU-UU--U---CGAA---CGAUACUCCGGGUAUAGAUAGAUGGUGAUAUCGCUAAUAAAGCGACGUCGUUGGAAAAGCUGCAGAAGAUCGCAGU ((((((((.........(((-((--(---(.((---((((.(.(((..((....))..))).)...(((((.....))))).)))))))))))))......))))))))... ( -26.26) >DroSim_CAF1 106116 103 + 1 GUGAUUUUAUCAUUAAUCUU-CU--U---CGAA---CGAUACUCCGGGUACAGAUAGAUGGUGAUGUCGCUGAUAAAGCGACGUCGUUGGAAAAGCUGUAGAAGAUCGCAGU ((((((((............-..--.---....---((......))..(((((.....(..((((((((((.....))))))))))..)......))))).))))))))... ( -30.90) >DroEre_CAF1 131175 103 + 1 GUGAUUUUGUCGUUAAUCUU-UU--U---CGAC---CGGUACUCCGGAUACAGAUAGAUGGUGAUGUCGCUUAUAAAGCGGCGUCGCUGGAAAAGCUGCAGCAGAUCGCAGU (((((((.((.((....(((-((--(---(..(---(((....))))............((((((((((((.....)))))))))))))))))))..)).)))))))))... ( -39.60) >DroYak_CAF1 105030 103 + 1 GUGAUUUUAUCAUUGAUCUU-UU--U---CGAC---CGGUACUGCGGAUACAGAUAGAUGGUGAUGUCGCUGAUGAAGCGACGUCGCUGGAAAAGCUGUAGAAGAUCGCAGU ((((((((....((((....-..--)---)))(---((......))).(((((.....(((((((((((((.....)))))))))))))......))))).))))))))... ( -37.80) >consensus GUGAUUUUAUCAUUAAUCUU_UU__U___CGAA___CGAUACUCCGGGUACAGAUAGAUGGUGAUGUCGCUGAUAAAGCGACGUCGCUGGAAAAGCUGCAGAAGAUCGCAGU ((((((((............................((......))....(((.....(((((((((((((.....)))))))))))))......)))...))))))))... (-20.57 = -21.18 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:04 2006