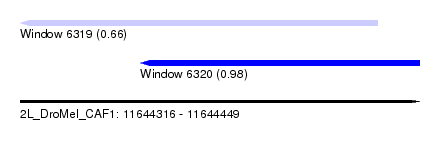
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,644,316 – 11,644,449 |
| Length | 133 |
| Max. P | 0.975094 |

| Location | 11,644,316 – 11,644,435 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.36 |
| Mean single sequence MFE | -32.87 |
| Consensus MFE | -21.08 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
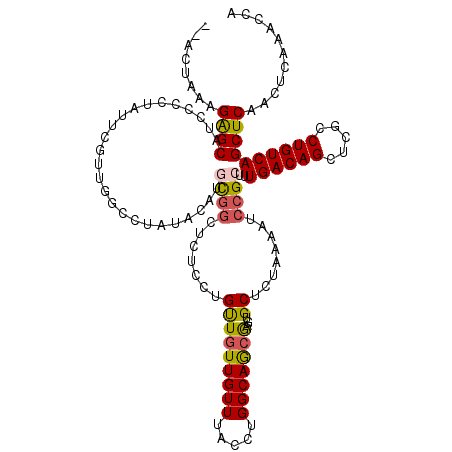
>2L_DroMel_CAF1 11644316 119 - 22407834 A-ACUAAAGGGCAUCCCAUAUUCUUUGGCCUAUAUACGUGGCUUUUCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAAAUCCGCUUUGACAGCUCGGCUGUCAGCUCAACUCAAACCA .-.....(((((((............((((.........))))......((((((((.....))))))))))))))).......(((..((((((...)))))))))............. ( -29.40) >DroPse_CAF1 91496 118 - 1 AAUCGAAAGAGCAUCUUUGGGUCGUUGUGCCGUGC-CGCGGC-GCCCUGUUGUUGUUUACCUGGCAGCGAGUGCUCCAAAAUCCGCUUUGACAGCUCGCCUGUCAGCUCAACUCAAACCG ..((....))........(((.(((((((......-))))))-))))((..((((.....((((((((((((....((((......))))...))))).)))))))..)))).))..... ( -37.60) >DroSec_CAF1 97695 117 - 1 A-ACUAUAGGGCAUCCCCUAUUCUUUGGCCUAUUC-UUUGGC-CUCCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAAAUCCGCUUUGACAGCUCGGCUGUCAGCUCAACUCAAACCA .-....((((((((............((((.....-...)))-).....((((((((.....))))))))))))))))......(((..((((((...)))))))))............. ( -34.20) >DroEre_CAF1 127530 118 - 1 --AAUGCAGAGCAUCCGCUAUUCGUUGGCAUAUAUAUGCGCUGCCUCUGUUGUUGUUUACCUGGCAGCAAGUGCUCUAAAAUCCGCUUUGACAGCUCGGCUGUCAGCUCAACUCAAACCA --...(((((((((..((((.....))))..........((((((...((........))..))))))..)))))))...........(((((((...)))))))))............. ( -36.40) >DroYak_CAF1 101369 113 - 1 -------AGAGCAUCCCCCAUUCGUUGGCAUAUAUAAGCGGCUCUUCCGCUGUUGUUUACCUGGCAGCAAGUGCUCUAAAAUCCGCUUUGACAGCUCGCCUGUCAGCUCAACUCAAACCA -------.((((..............(((((.....(((((.....)))))((((((.....))))))..))))).............((((((.....))))))))))........... ( -30.00) >DroAna_CAF1 111529 115 - 1 -----UAAGAGCAUUUGGCAUUCGUUAUCCUGGCCAUCCUGAUGUCCUGUUUUUGUUUACCUGGCAACGAGUGCUCUAAAAUCCGCUUUGACAGCUCGCCUGUCAGCUCAACUCAAACCA -----...((((....((((((((((.....((.(((....))).)).......(((.....))))))))))))).............((((((.....))))))))))........... ( -29.60) >consensus __ACUAAAGAGCAUCCCCUAUUCGUUGGCCUAUACAUGCGGCUCUCCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAAAUCCGCUUUGACAGCUCGCCUGUCAGCUCAACUCAAACCA ........((((.........................((((.......(((((((((.....)))))))...))........))))..((((((.....))))))))))........... (-21.08 = -20.98 + -0.11)


| Location | 11,644,356 – 11,644,449 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 74.08 |
| Mean single sequence MFE | -22.27 |
| Consensus MFE | -13.84 |
| Energy contribution | -13.62 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11644356 93 - 22407834 AAACAAUUUUCACCAACUAAAGGGCAUCCCAUAUUCUUUGGCCUAUAUACGUGGCUUUUCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAA ....................(((((((............((((.........))))......((((((((.....)))))))))))))))... ( -19.20) >DroSec_CAF1 97735 91 - 1 AAAGAAUUUUUACCAACUAUAGGGCAUCCCCUAUUCUUUGGCCUAUUC-UUUGGC-CUCCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAA (((((((.....((((..((((((....))))))...))))...))))-)))(((-((((((((...........))))).))).)))..... ( -24.80) >DroSim_CAF1 102389 91 - 1 AAAGAAUUUUUAC--ACUAUAGAGCAUCCCCUAUUCUUUGGCCUAUAUAUGCGGCUCUCCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAA .............--....((((((((............((((.........))))......((((((((.....)))))))))))))))).. ( -20.90) >DroEre_CAF1 127570 89 - 1 UAACCAAUUUU----AAUGCAGAGCAUCCGCUAUUCGUUGGCAUAUAUAUGCGCUGCCUCUGUUGUUGUUUACCUGGCAGCAAGUGCUCUAAA ...........----.....(((((((..((((.....))))..........((((((...((........))..))))))..)))))))... ( -25.40) >DroYak_CAF1 101409 84 - 1 UUAAAAGUUUU---------AGAGCAUCCCCCAUUCGUUGGCAUAUAUAAGCGGCUCUUCCGCUGUUGUUUACCUGGCAGCAAGUGCUCUAAA ........(((---------(((((((...(((.....)))........(((((.....)))))((((((.....))))))..)))))))))) ( -24.50) >DroAna_CAF1 111569 89 - 1 CACUUAUUUUCUUC----UAAGAGCAUUUGGCAUUCGUUAUCCUGGCCAUCCUGAUGUCCUGUUUUUGUUUACCUGGCAACGAGUGCUCUAAA ..............----..((((((((((..............((.(((....))).)).....(((((.....)))))))))))))))... ( -18.80) >consensus AAACAAUUUUUAC__ACUAAAGAGCAUCCCCUAUUCGUUGGCCUAUAUAUGCGGCUCUCCUGUUGUUGUUUACCUGGCAGCGAGUGCUCUAAA ....................(((((((............(((...........)))......((((((((.....)))))))))))))))... (-13.84 = -13.62 + -0.22)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:03 2006