
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,639,037 – 11,639,131 |
| Length | 94 |
| Max. P | 0.639705 |

| Location | 11,639,037 – 11,639,131 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.44 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.30 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11639037 94 - 22407834 CGUCACUUGUUUGACAUAUCUACAAGGCGCAAUCUGAUUGCGC-------C-GCAUA------UAUGUUUGUGUGCU----------UAAUGAUAUGUACA-AG-GACUGGCCAAACAUU .(((.(((((...(((((((.....((((((((...)))))))-------)-(((((------((....))))))).----------....))))))))))-))-)))............ ( -35.10) >DroPse_CAF1 87657 103 - 1 CGUCACUUGUCUGACAUAUCUACAAGGCGCAAUCUGAGGGGGGGUCACCCCUCCCCU------UGUGUGUGUGUGUU----------UAAUGAUAUGUACA-AAAGCUUGACCAAACAUU .(((((((.....(((((((.(((..(((((...((((((((((.....))))))))------))..))))).))).----------....)))))))...-.)))..))))........ ( -35.50) >DroSim_CAF1 96267 94 - 1 CGUCACUUGUUUGACAUAUCUACAAGGCGCAAUCUGAUUGCGC-------C-GCAUA------UAUGUUUGUGUGCU----------UAAUGAUAUGUACA-AG-GACUGGCCAAACAUU .(((.(((((...(((((((.....((((((((...)))))))-------)-(((((------((....))))))).----------....))))))))))-))-)))............ ( -35.10) >DroEre_CAF1 122482 94 - 1 CGUCACUUGUUUGACAUAUCUACAAGGCGCAAUCUGAUUGCGC-------C-GCACA------UUUGUUUGUGUGCU----------UAAUGAUAUGUACA-AG-GACUGGCCAAACAUU .(((.(((((...(((((((.....((((((((...)))))))-------)-(((((------(......)))))).----------....))))))))))-))-)))............ ( -35.90) >DroYak_CAF1 95883 110 - 1 CGUCACUUGUUUGACAUAUCUACAAGGCGCAAUCUGAUUGCGC-------C-GCAUAAACGAGUAUGUUUGUGUGAUAUGUAACAACUAAUGAUAUGUACA-AG-GACUGGCCAAACAUU .(((.(((((...(((((((.....((((((((...)))))))-------)-((((((((......)))))))).................))))))))))-))-)))............ ( -35.00) >DroAna_CAF1 105923 86 - 1 CGUCACUUGUUUGACAUAUCUACGAGGCGAAAUCAGAUUG-----------------------UACGUUUGUGUGCU----------UAAUGAUAUGUACAAAG-GCCUGCCCAAACAUU .......(((((((((((((....(((((....(((((..-----------------------...)))))..))))----------)...))))))).....(-(....)))))))).. ( -21.10) >consensus CGUCACUUGUUUGACAUAUCUACAAGGCGCAAUCUGAUUGCGC_______C_GCAUA______UAUGUUUGUGUGCU__________UAAUGAUAUGUACA_AG_GACUGGCCAAACAUU .((((.((...(((((((((.....((((((.......(((...........)))................))))))..............))))))).))....)).))))........ ( -9.90 = -10.44 + 0.54)
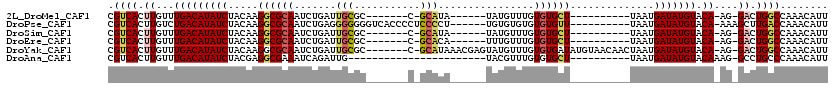

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:55 2006