
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,637,376 – 11,637,482 |
| Length | 106 |
| Max. P | 0.904948 |

| Location | 11,637,376 – 11,637,482 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 75.08 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -13.45 |
| Energy contribution | -14.15 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11637376 106 + 22407834 -GAUAAGCCUGAGCCAGAACAUAAUUCUUGAGUAUUUUCUGUGAAUUAGCUGG-----CAUACUCCAGCCAUAAACAGGCUCGCA---UCCUGCUCAUCCUCUCGCCAACU--CAAU -(((.(((..(((((((....((((((..(((....)))...)))))).))))-----).......((((.......))))....---))..))).)))............--.... ( -24.60) >DroPse_CAF1 85541 92 + 1 -GAUAAGCCAGAGCCAACGCAUAAUUCUUGAGUAUUUUCUGUGAAUUAGCUGG-----CAUUUCUAUGCCAUAAACAGGGUCCU-GGAUCUG------UUUUCUG------------ -((((..((((..((...((.((((((..(((....)))...))))))))(((-----(((....)))))).......))..))-))...))------)).....------------ ( -26.10) >DroSim_CAF1 94490 106 + 1 -GAUAAGCCUGAUCCAGCACAUAAUUCUUGAGUAUUUUCUGUGAAUUAGCUGG-----CAUACUCCAGCCAUAAACAGGCUCGCA---UCCUGCUCUUCCUCUCGCCAACU--CAAU -....((((((..........((((((..(((....)))...))))))(((((-----......)))))......)))))).((.---....)).................--.... ( -22.80) >DroEre_CAF1 120727 106 + 1 -GAUAAGCCUGAGCCAGCACAUAAUUCUUGAGUAUUUUCUGUGAAUUAGCUGG-----CAUACUCGAGCCAUAAACAGGCUCGCA---UCCUGCUCAUCCUCUCGCCAACU--CAAU -(((.(((..((((((((...((((((..(((....)))...)))))))))))-----).....((((((.......))))))..---))..))).)))............--.... ( -31.50) >DroYak_CAF1 94202 95 + 1 -GAUAAGCCUGAGCCAGCACAUAAUUCUUGAGUAUUUUCUGUGAAUUAGCUGG-----CAUACACUAGCCAUAAACAGGCUCGC----------------UCUUGCCAACUCUCCAU -....((((((.((((((...((((((..(((....)))...)))))))))))-----)......((....))..)))))).((----------------....))........... ( -24.70) >DroAna_CAF1 104419 106 + 1 GGAUAAGCCUGAGCCAGCACAUAAUUCUUGAGUAUUUUCUGGAAACUAGCUGGGAUCGCAUCUCUUAGUCAUAAACAGGCUGCCAGGGUCCGAAUUAUUUUCCUGC----------- ((...((((((..(((((......((((.(((....))).))))....)))))(((...........))).....)))))).))((((............))))..----------- ( -25.70) >consensus _GAUAAGCCUGAGCCAGCACAUAAUUCUUGAGUAUUUUCUGUGAAUUAGCUGG_____CAUACUCCAGCCAUAAACAGGCUCGCA___UCCUGCUCAUCCUCUCGCCAACU__CAAU .....((((((..........((((((..(((....)))...))))))(((((...........)))))......)))))).................................... (-13.45 = -14.15 + 0.70)

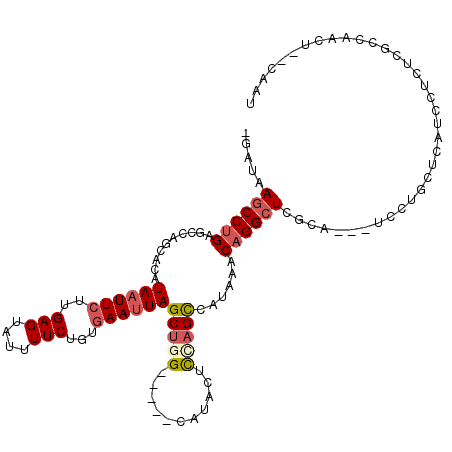
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:53 2006