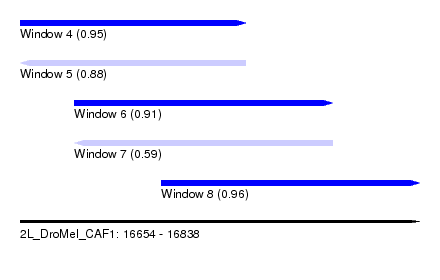
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 16,654 – 16,838 |
| Length | 184 |
| Max. P | 0.962505 |

| Location | 16,654 – 16,758 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -24.82 |
| Energy contribution | -27.93 |
| Covariance contribution | 3.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16654 104 + 22407834 UCAACAAA-GCUUCAGGUACCUCCUCAUAUUUGGCACCGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAACCUUUUU-UAUGCGGUGCUUUGU ........-((((...((((.((((..(((.((....)).)))..))))))))...........))))...(((((((((((........-..))))))))))).. ( -28.44) >DroSec_CAF1 4811 104 + 1 -CCGCAAUGUCUGUGCGUACCUGCCCAUAUUUGGCACAGCAUAGAAGGAGAGCACAAAAU-AACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUUUGU -..((....(((((((.....((((.......))))..)))))))......)).......-..........((((((((((((((.....)))))))))))))).. ( -37.50) >DroSim_CAF1 4916 105 + 1 -CCGCAAUGUCUGUGCGUACCUGCGCAUAUUUGGCACAGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUGUGU -..(((...(((((((.....(((.((....)))))..))))))).....))).............(((....((((((((((((.....)))))))))))).))) ( -38.70) >consensus _CCGCAAUGUCUGUGCGUACCUGCCCAUAUUUGGCACAGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUUUGU ...((......((((((......))))))((((((((..(......)..)))).))))........))...((((((((((((((.....)))))))))))))).. (-24.82 = -27.93 + 3.11)

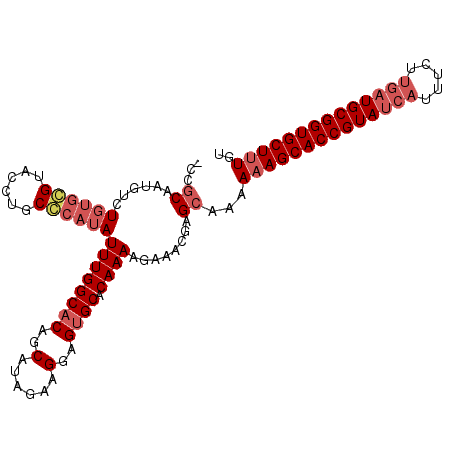
| Location | 16,654 – 16,758 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 85.80 |
| Mean single sequence MFE | -33.17 |
| Consensus MFE | -25.45 |
| Energy contribution | -27.90 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16654 104 - 22407834 ACAAAGCACCGCAUA-AAAAAGGUUACGGUGCUUUUUUGCUCGUUUCUUUUGUGCACUCCUUCUAUGCGGUGCCAAAUAUGAGGAGGUACCUGAAGC-UUUGUUGA ((((((((((.....-.....)))...((((((((....(((((....((((.(((((.(......).))))))))).)))))))))))))....))-)))))... ( -32.70) >DroSec_CAF1 4811 104 - 1 ACAAAGCACCGCAUCAAGAAAUGAUACGGUGCUUUUUUGCUCGUU-AUUUUGUGCUCUCCUUCUAUGCUGUGCCAAAUAUGGGCAGGUACGCACAGACAUUGCGG- ..(((((((((.((((.....)))).)))))))))(((((((((.-..((((.((.(..(......)..).)))))).)))))))))..((((.......)))).- ( -34.40) >DroSim_CAF1 4916 105 - 1 ACACAGCACCGCAUCAAGAAAUGAUACGGUGCUUUUUUGCUCGUUUCUUUUGUGCACUCCUUCUAUGCUGUGCCAAAUAUGCGCAGGUACGCACAGACAUUGCGG- ....(((((((.((((.....)))).)))))))..(((((.(((....((((.((((..(......)..)))))))).))).)))))..((((.......)))).- ( -32.40) >consensus ACAAAGCACCGCAUCAAGAAAUGAUACGGUGCUUUUUUGCUCGUUUCUUUUGUGCACUCCUUCUAUGCUGUGCCAAAUAUGAGCAGGUACGCACAGACAUUGCGG_ ..(((((((((.((((.....)))).)))))))))(((((((((....((((.((((..(......)..)))))))).)))))))))..((((.......)))).. (-25.45 = -27.90 + 2.45)

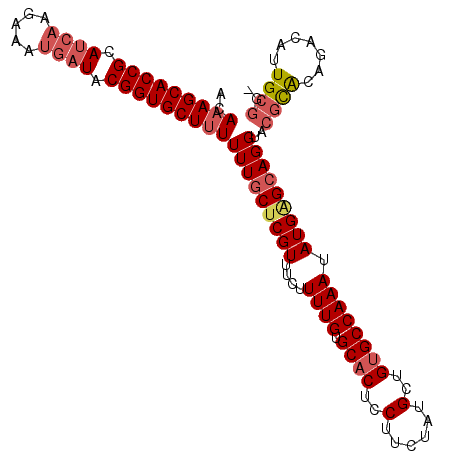
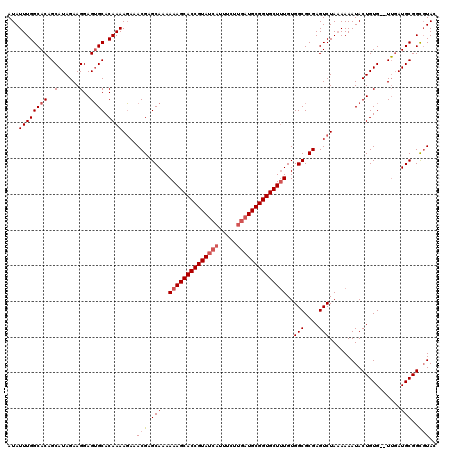
| Location | 16,679 – 16,798 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -31.89 |
| Energy contribution | -33.67 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.99 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16679 119 + 22407834 AUAUUUGGCACCGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAACCUUUUU-UAUGCGGUGCUUUGUGGCGCGAGUCUACAAAAUACUGUGUGUGUAUGCGGUUUAC .........((((((((.(.(.((((......(((..((.((...(((((((((((........-..)))))))))))...)).))..)))......)))).)...).)))))))).... ( -39.40) >DroSec_CAF1 4836 117 + 1 AUAUUUGGCACAGCAUAGAAGGAGAGCACAAAAU-AACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUUUGUGGCGCGAGUCUAAAAAAUACUGUG--UUGAUGCGGCGUAC .(((.(.(((((((((((..(.......).....-..((.((...((((((((((((((.....))))))))))))))...)).)).............)))))--))).))).).))). ( -42.30) >DroSim_CAF1 4941 118 + 1 AUAUUUGGCACAGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUGUGUGGCGCGAGUCUAAAAAAUACUGUG--UUGAUGCGGCGUAC .(((.(.(((((((((((..(.......)...(((..((.((.....((((((((((((.....)))))))))))).....)).))..)))........)))))--))).))).).))). ( -42.50) >consensus AUAUUUGGCACAGCAUAGAAGGAGUGCACAAAAGAAACGAGCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUUUGUGGCGCGAGUCUAAAAAAUACUGUG__UUGAUGCGGCGUAC ...((((((((..(......)..)))).))))....(((.(((..((((((((((((((.....))))))))))))))..(((....)))....................)))..))).. (-31.89 = -33.67 + 1.78)

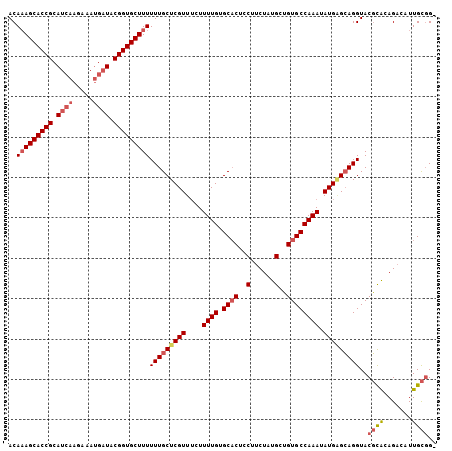
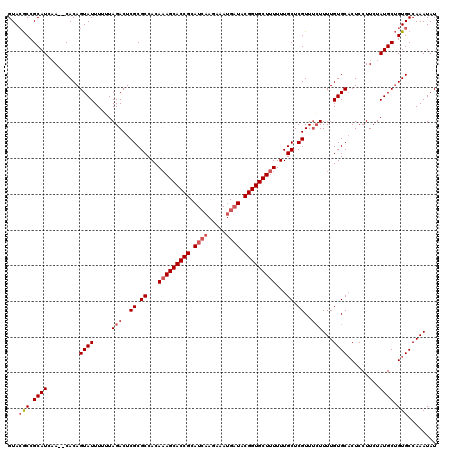
| Location | 16,679 – 16,798 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -23.99 |
| Energy contribution | -26.10 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16679 119 - 22407834 GUAAACCGCAUACACACACAGUAUUUUGUAGACUCGCGCCACAAAGCACCGCAUA-AAAAAGGUUACGGUGCUUUUUUGCUCGUUUCUUUUGUGCACUCCUUCUAUGCGGUGCCAAAUAU ....((((((((........((((.....(((..((.((...(((((((((....-..........)))))))))...)).))..)))...))))........))))))))......... ( -32.53) >DroSec_CAF1 4836 117 - 1 GUACGCCGCAUCAA--CACAGUAUUUUUUAGACUCGCGCCACAAAGCACCGCAUCAAGAAAUGAUACGGUGCUUUUUUGCUCGUU-AUUUUGUGCUCUCCUUCUAUGCUGUGCCAAAUAU (((((..((((...--...(((((..........((.((...(((((((((.((((.....)))).)))))))))...)).))..-.....)))))........)))))))))....... ( -28.67) >DroSim_CAF1 4941 118 - 1 GUACGCCGCAUCAA--CACAGUAUUUUUUAGACUCGCGCCACACAGCACCGCAUCAAGAAAUGAUACGGUGCUUUUUUGCUCGUUUCUUUUGUGCACUCCUUCUAUGCUGUGCCAAAUAU (((((..((((...--....((((.....(((..((.((.....(((((((.((((.....)))).))))))).....)).))..)))...)))).........)))))))))....... ( -29.09) >consensus GUACGCCGCAUCAA__CACAGUAUUUUUUAGACUCGCGCCACAAAGCACCGCAUCAAGAAAUGAUACGGUGCUUUUUUGCUCGUUUCUUUUGUGCACUCCUUCUAUGCUGUGCCAAAUAU ...(((.((((.........((((.....(((..((.((...(((((((((.((((.....)))).)))))))))...)).))..)))...)))).........)))).)))........ (-23.99 = -26.10 + 2.11)


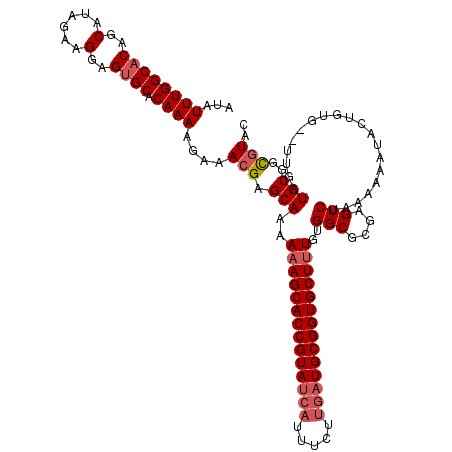
| Location | 16,719 – 16,838 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -34.08 |
| Energy contribution | -35.41 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 16719 119 + 22407834 GCAAAAAAGCACCGUAACCUUUUU-UAUGCGGUGCUUUGUGGCGCGAGUCUACAAAAUACUGUGUGUGUAUGCGGUUUACUUGUAGUUGCGGUGGAAAAAAUUUACCUAAAUGUUAAUGC ((...(((((((((((........-..)))))))))))...))((((..((((((...(((((((....)))))))....))))))))))((((((.....))))))............. ( -37.60) >DroSec_CAF1 4875 118 + 1 GCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUUUGUGGCGCGAGUCUAAAAAAUACUGUG--UUGAUGCGGCGUACUUGUAGUUGCGGUGGACAAAAUUUACCUAAAUGUAAAUGC ((...((((((((((((((.....))))))))))))))...))((..(((((.......((((.--.....))))((((........)))).)))))...((((((......)))))))) ( -39.30) >DroSim_CAF1 4981 118 + 1 GCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUGUGUGGCGCGAGUCUAAAAAAUACUGUG--UUGAUGCGGCGUACUUGUAGUUGCGGUGGACAAAAUUUACCUAAAUGUAAAUGC (((....((((((((((((.....)))))))))))).)))(((....)))............((--((.(((((((.........))))).)).))))..((((((......)))))).. ( -38.10) >consensus GCAAAAAAGCACCGUAUCAUUUCUUGAUGCGGUGCUUUGUGGCGCGAGUCUAAAAAAUACUGUG__UUGAUGCGGCGUACUUGUAGUUGCGGUGGACAAAAUUUACCUAAAUGUAAAUGC (((..((((((((((((((.....))))))))))))))(..((((((((.......(..((((........))))..))))))).))..)((((((.....))))))..........))) (-34.08 = -35.41 + 1.33)

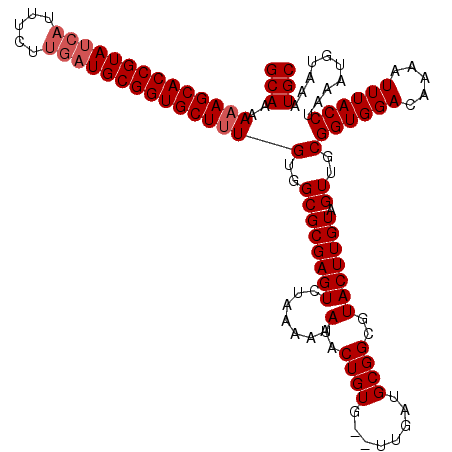
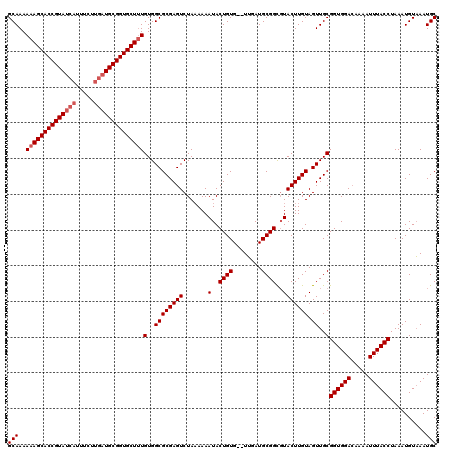
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:03 2006