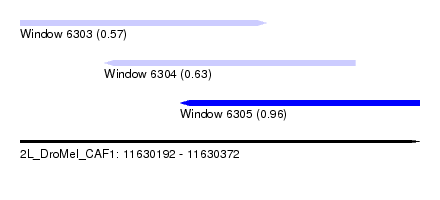
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,630,192 – 11,630,372 |
| Length | 180 |
| Max. P | 0.961327 |

| Location | 11,630,192 – 11,630,303 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.80 |
| Mean single sequence MFE | -29.08 |
| Consensus MFE | -16.86 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11630192 111 + 22407834 AAUUAGAAUAAUUUGGCCAGCUGUUGCUACAACUCAGCUCCUCCUCCACCUCAGAUGCUG-----CUCCUUGGUCAUUCUGC-UGGUGAUUACACCAGGAUAUUUGCCAAAAUAUUU .....(((((.(((((((((..((.((((.....((((..((..........))..))))-----.....)))).)).)))(-(((((....)))))).......)))))).))))) ( -26.20) >DroPse_CAF1 78075 97 + 1 AAUUAGAAUAAUUUUGUCAGCUGCUGCUAGAGCCGAGGCCCC-------------UGCUG-----CUCUGUGGCUG--GUGAUGGGUGAUGGGACUUAAAUAUUUGCCAAAAUAUUU ............((..(((.(.((..(.(((((.(((....)-------------).).)-----)))))..)).)--.)))..))...........((((((((....)))))))) ( -24.70) >DroSec_CAF1 83523 111 + 1 AAUUAGAAUAAUUUGGCCAGCUGUUGCUACAACUCAGCUCCUCCUCCUCUUCAUAUGCUG-----CUCCUUGGCCAUUCUGC-UGGUGAUUACACCAGGAUAUUUGCCAAAAUAUUU ...(((((.....((((((((....)))......((((..................))))-----......))))))))))(-(((((....))))))(((((((....))))))). ( -27.37) >DroSim_CAF1 87313 111 + 1 AAUUAGAAUAAUUUGGCCAGCUGUUGCUACAACUCAGCUCCUCCUCCUCUUCGGAUGCUG-----CUCGUUGGCCAUUCUGC-UGGUGAUUACACCAGGAUAUUUGCCAAAAUAUUU ...(((((.....((((((((.((((...)))).(((((((...........))).))))-----...)))))))))))))(-(((((....))))))(((((((....))))))). ( -34.60) >DroEre_CAF1 113825 108 + 1 AAUUAGAAUAAUUUGGGCAGCUGUUGCUACAACACAGCUCCUCC---UCUUCAAAUGCUG-----CUCCUUGGCCAUUCUGC-UGGUGAUUACACCAGGAUAUUUGCCAAAAUAUUU .....(((((....(((((((.((((........))))......---.........))))-----))).(((((.....(.(-(((((....)))))).).....)))))..))))) ( -27.09) >DroYak_CAF1 86976 113 + 1 AAUUAGAAUAAUUUGGCCAGCUGUUGCUACAACUCAGCUCCUUC---UCUGCAGGUGCAGCUCAGCUCCUUGGCCAUUCUGC-UGGUGAUUACACCAGGAUAUUUGCCAAAAUAUUU ...(((((.....((((((((((..(((.((.(((((.......---.))).)).)).))).))))....)))))))))))(-(((((....))))))(((((((....))))))). ( -34.50) >consensus AAUUAGAAUAAUUUGGCCAGCUGUUGCUACAACUCAGCUCCUCC___UCUUCAGAUGCUG_____CUCCUUGGCCAUUCUGC_UGGUGAUUACACCAGGAUAUUUGCCAAAAUAUUU ...(((((.....((((((((....)))......((((..................))))...........))))))))))..(((((....)))))((((((((....)))))))) (-16.86 = -17.29 + 0.42)

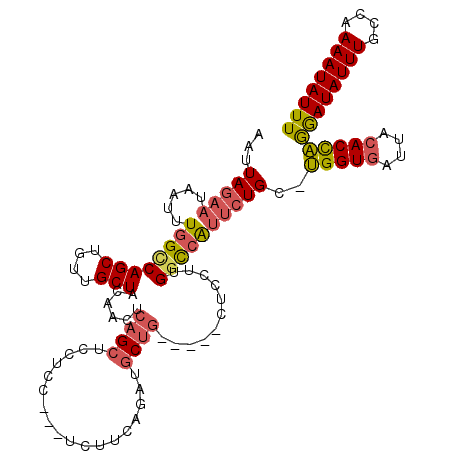
| Location | 11,630,230 – 11,630,343 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 91.02 |
| Mean single sequence MFE | -29.73 |
| Consensus MFE | -24.12 |
| Energy contribution | -25.16 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11630230 113 - 22407834 CUAUUAACUUCUCGAGUUCACAGUCCAAAUUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGAAUGACCAAGGAG-----CAGCAUCUGAGGUGGAGGAGGA .......((((((..(.(((((((....))))))).)......(((((((.........((((((....)))))))))))))(((..(((.-----.....)))..))).)))))).. ( -29.90) >DroSec_CAF1 83561 113 - 1 CUAUUUACUUCUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGAAUGGCCAAGGAG-----CAGCAUAUGAAGAGGAGGAGGA .......((((((..(.(((((.(....).))))).)........(((((((.......((((((....)))))).......)))))))..-----...........))))))..... ( -30.44) >DroSim_CAF1 87351 113 - 1 CUAUUUACUUCUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGAAUGGCCAACGAG-----CAGCAUCCGAAGAGGAGGAGGA .......((.((((.(.(((((.(....).))))).)..........(((((.......((((((....)))))).......)))))))))-----.))..(((.....)))...... ( -29.14) >DroEre_CAF1 113863 110 - 1 CUAUUUACUAUUUGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGAAUGGCCAAGGAG-----CAGCAUUUGAAGA---GGAGGA ((.((((.(((((((..(((((.(....).)))))...)))))))(((((((.......((((((....)))))).......)))))))..-----.......)))).)---)..... ( -25.74) >DroYak_CAF1 87014 115 - 1 CUAUUUACUUUUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGAAUGGCCAAGGAGCUGAGCUGCACCUGCAGA---GAAGGA .......((((((.((((((...(((..((((....)))).......(((((.......((((((....)))))).......))))))))..))))))((....)).))---)))).. ( -33.44) >consensus CUAUUUACUUCUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGAAUGGCCAAGGAG_____CAGCAUCUGAAGAGGAGGAGGA .......((((((..(.(((((.(....).))))).)........(((((((.......((((((....)))))).......))))))).....................)))))).. (-24.12 = -25.16 + 1.04)

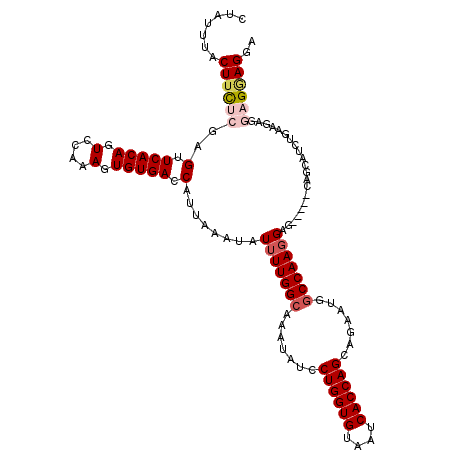
| Location | 11,630,264 – 11,630,372 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -18.47 |
| Consensus MFE | -17.08 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 11630264 108 - 22407834 AUUUAAUCAAUCCUCUUAGCCAUCCCAUCCUAUUAACUUCUCGAGUUCACAGUCCAAAUUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGA ..................((((.............((((...))))(((((((....))))))).............)))).......((((((....)))))).... ( -19.70) >DroSec_CAF1 83595 108 - 1 AUUUAAUCAAUCCUCUUAGCUAUCCCAUCCUAUUUACUUCUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGA ..................(((..............................(.(((((((((.........)))))))))).........((((....)))))))... ( -16.60) >DroSim_CAF1 87385 108 - 1 AUAUAAUCAAUCCUCUUAGCCAUCCCAUCCUAUUUACUUCUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGA ..................((((.............((((...))))(((((.(....).))))).............)))).......((((((....)))))).... ( -18.00) >DroEre_CAF1 113894 108 - 1 AUUCAAUCAAUCAUCGCAGCCAUCCUAUCCUAUUUACUAUUUGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGA ...............((.((((...............(((((((..(((((.(....).)))))...)))))))...))))........(((((....)))))))... ( -20.27) >DroYak_CAF1 87050 108 - 1 AUUUAAUCAAUCUUCUUGGCCAUCCCAUCCUAUUUACUUUUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGA ..................((((.............((((...))))(((((.(....).))))).............)))).......((((((....)))))).... ( -17.80) >consensus AUUUAAUCAAUCCUCUUAGCCAUCCCAUCCUAUUUACUUCUCGAGUUCACAGUCCAAAGUGUGACCAUUAAAUAUUUUGGCAAAUAUCCUGGUGUAAUCACCAGCAGA ..................((((.............(((.....)))(((((.(....).))))).............)))).......((((((....)))))).... (-17.08 = -16.92 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:48 2006