| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,627,927 – 11,628,022 |
| Length | 95 |
| Max. P | 0.899516 |

| Location | 11,627,927 – 11,628,022 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -20.00 |
| Energy contribution | -21.45 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
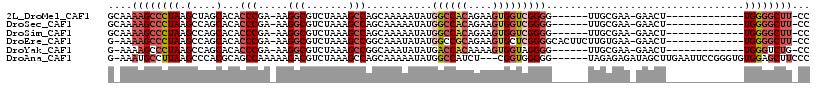
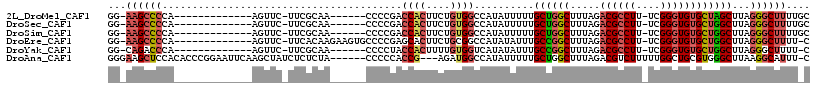
>2L_DroMel_CAF1 11627927 95 + 22407834 GCAAAAGCCCUAAGCUAGCACACCCGA-AAGGCGUCUAAAGCCAGCAAAAAUAUGGCCACAGAAGUGGUCGGGG------UUGCGAA-GAACU-------------UGGGGCUU-CC ....((((((((((((.(((.((((..-..(((.......)))..........(((((((....))))))))))------))))..)-)..))-------------))))))))-.. ( -39.40) >DroSec_CAF1 81279 95 + 1 GCAAAAGCCCUAAGCCAGCACACCCGA-AAGGCGUCUAAAGCCAGCAAAAAUAUGGCCACAGAAGUGGUCGGGG------UUGCGAA-GAACU-------------UGGGGCUU-CC ....((((((((((...(((.((((..-..(((.......)))..........(((((((....))))))))))------))))...-...))-------------))))))))-.. ( -39.10) >DroSim_CAF1 85051 95 + 1 GCAAAAGCCCUAAGCCAGCACACCCGA-AAGGCGUCUAAAGCCAGCAAAAAUAUGGCCACAGAAGUGGUCGGGG------UUGCGAA-GAACU-------------UGGGGCUU-CC ....((((((((((...(((.((((..-..(((.......)))..........(((((((....))))))))))------))))...-...))-------------))))))))-.. ( -39.10) >DroEre_CAF1 111558 100 + 1 G-AAAAGCCCUAAGCCAGCACACCCGA-AAGGCGUCUAAAGCCGGCAAAUAUAUGGCCGCAGAAGUGCUCGGGGCACUUCUUGUGAA-GAACU-------------UGGGGCUU-CC .-..((((((((((....(((((((..-..)).)).....((.(((.........)))))((((((((.....)))))))).)))..-...))-------------))))))))-.. ( -39.60) >DroYak_CAF1 84683 94 + 1 G-AAAAGCCCUAAGCCAGCACACCCGA-AAGGCGUCUAAAGCCGGCAAAUAUAUGACCACAAAAGUGGUAGGGG------UUGCGAA-GAACU-------------UGGGUCUG-CC .-...((.((((((...(((..(((..-..(((.......)))............(((((....))))).))).------.)))...-...))-------------)))).)).-.. ( -28.60) >DroAna_CAF1 94687 107 + 1 G-AAAUGCCUUAAGCCCACGCAGCCAAAAAGACGUCUAAAGCCAGCAAAAAUAUGGCCAUCU---CGGUGGGGG------UAGAGAGAUAGCUUGAAUUCCGGGUGUGGAGCUUCCC .-.........((((((((((.(((....((....))...((((.........)))).....---.))).((((------(.(((......)))..)))))..)))))).))))... ( -26.80) >consensus G_AAAAGCCCUAAGCCAGCACACCCGA_AAGGCGUCUAAAGCCAGCAAAAAUAUGGCCACAGAAGUGGUCGGGG______UUGCGAA_GAACU_____________UGGGGCUU_CC ....((((((((.(....)...(((.....(((.......)))...........((((((....))))))))).................................))))))))... (-20.00 = -21.45 + 1.45)
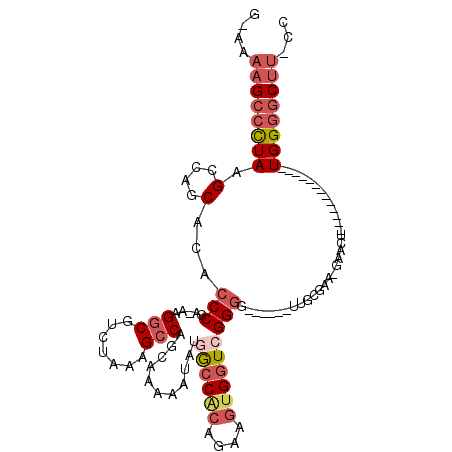
| Location | 11,627,927 – 11,628,022 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -20.90 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
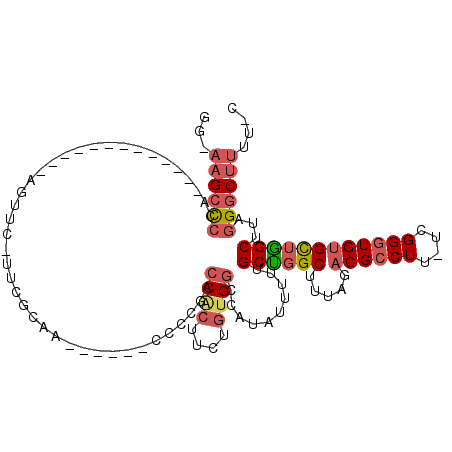
>2L_DroMel_CAF1 11627927 95 - 22407834 GG-AAGCCCCA-------------AGUUC-UUCGCAA------CCCCGACCACUUCUGUGGCCAUAUUUUUGCUGGCUUUAGACGCCUU-UCGGGUGUGCUAGCUUAGGGCUUUUGC ((-((((((.(-------------((((.-...(((.------.(((((...(.((((.(((((((....)).))))).)))).)....-)))))..))).))))).)))))))).. ( -36.50) >DroSec_CAF1 81279 95 - 1 GG-AAGCCCCA-------------AGUUC-UUCGCAA------CCCCGACCACUUCUGUGGCCAUAUUUUUGCUGGCUUUAGACGCCUU-UCGGGUGUGCUGGCUUAGGGCUUUUGC ((-((((((.(-------------((((.-...(((.------.(((((...(.((((.(((((((....)).))))).)))).)....-)))))..))).))))).)))))))).. ( -36.40) >DroSim_CAF1 85051 95 - 1 GG-AAGCCCCA-------------AGUUC-UUCGCAA------CCCCGACCACUUCUGUGGCCAUAUUUUUGCUGGCUUUAGACGCCUU-UCGGGUGUGCUGGCUUAGGGCUUUUGC ((-((((((.(-------------((((.-...(((.------.(((((...(.((((.(((((((....)).))))).)))).)....-)))))..))).))))).)))))))).. ( -36.40) >DroEre_CAF1 111558 100 - 1 GG-AAGCCCCA-------------AGUUC-UUCACAAGAAGUGCCCCGAGCACUUCUGCGGCCAUAUAUUUGCCGGCUUUAGACGCCUU-UCGGGUGUGCUGGCUUAGGGCUUUU-C ((-((((((..-------------.....-......((((((((.....))))))))..............((((((.....(((((..-...)))))))))))...))))))))-. ( -37.80) >DroYak_CAF1 84683 94 - 1 GG-CAGACCCA-------------AGUUC-UUCGCAA------CCCCUACCACUUUUGUGGUCAUAUAUUUGCCGGCUUUAGACGCCUU-UCGGGUGUGCUGGCUUAGGGCUUUU-C ((-((.((((.-------------.....-...((((------.....(((((....))))).......)))).(((.......)))..-..)))).))))(((.....)))...-. ( -28.20) >DroAna_CAF1 94687 107 - 1 GGGAAGCUCCACACCCGGAAUUCAAGCUAUCUCUCUA------CCCCCACCG---AGAUGGCCAUAUUUUUGCUGGCUUUAGACGUCUUUUUGGCUGCGUGGGCUUAAGGCAUUU-C ...((((.((((.............((((((((....------........)---))))))).........((.((((..((....))....)))))))))))))).........-. ( -27.60) >consensus GG_AAGCCCCA_____________AGUUC_UUCGCAA______CCCCGACCACUUCUGUGGCCAUAUUUUUGCUGGCUUUAGACGCCUU_UCGGGUGUGCUGGCUUAGGGCUUUU_C ...((((((........................................((((....))))..........((((((.....((((((....))))))))))))...)))))).... (-20.90 = -21.82 + 0.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:44 2006