
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 11,626,908 – 11,627,055 |
| Length | 147 |
| Max. P | 0.604766 |

| Location | 11,626,908 – 11,627,026 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -21.26 |
| Consensus MFE | -13.54 |
| Energy contribution | -14.23 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
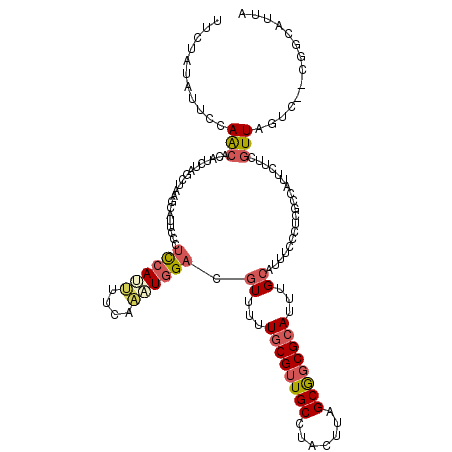
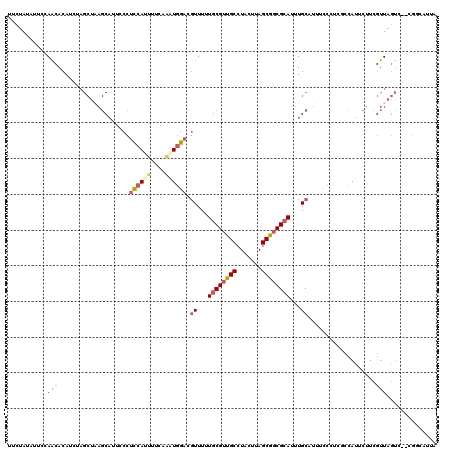
>2L_DroMel_CAF1 11626908 118 - 22407834 UGCUAUACUUCAACACAUCUAGCUAAGCAUUCCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAUUUGCAUUUCCCUCGCCAUUCUUCGUUAGUC--CGGCAUUA .((((..............))))...(((.....((((((....))))))......((((((((.......))))))))..))).........(((...((.....))..--.))).... ( -24.94) >DroSec_CAF1 80258 120 - 1 UCCUAUAUUCCAACACAUCUAGCUAAGCAUUCCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAUUUGCAUUUCCCUCUCCAUUCUUCGUUAGUCCCCGGCAUUA .........((.......(((((.(((.......((((((....)))))).((...((((((((.......))))))))...))...............))).))))).....))..... ( -23.00) >DroSim_CAF1 84030 118 - 1 UUCUAUAUUCCAACACAUCUAGCUAAGCAUUGCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAUUUGCAUUUCCCUCUCCAUUCUUCGUUAGUC--CGGCAUUA ..............................((((((((((....)))))).((...((((((((.......))))))))...))..........................--.))))... ( -23.90) >DroEre_CAF1 110571 110 - 1 UUCUGUGUUCCAACACAUCCAGCUUAGCAUUCCCUCCAGUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAUUUGCAUAUCCCUCUCCAUUCUUCGUUAGUC---------- ...(((((....)))))......(((((......((((........)))).((...((((((((.......))))))))...))...................)))))..---------- ( -22.80) >DroYak_CAF1 83654 110 - 1 UUUUAUAUUCCAACACAAAAAGCUAAGCAAUCCCUCCAGCUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAUUUGCAUUUUCCUCGCCAUUCUUCGUUAGUC---------- .....................(((((........((((........)))).((...((((((((.......))))))))...))....................))))).---------- ( -19.30) >DroAna_CAF1 93823 105 - 1 UUUAAUAUUCCAGCACAU------------U-CCAUCAACUUCAGUUAGAAUUUUUUCCGCUGCCUACUUAGCAACGCAUUUGCAUUUCCCAUGCCAGUCGUCCUUAUCC--CGGCUUUA ...........(((....------------.-.....(((....))).(((....))).)))(((....(((..(((.(((.((((.....)))).))))))..)))...--.))).... ( -13.60) >consensus UUCUAUAUUCCAACACAUCUAGCUAAGCAUUCCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAUUUGCAUUUCCCUCGCCAUUCUUCGUUAGUC__CGGCAUUA ...........(((....................((((((....)))))).((...((((((((.......))))))))...))...................))).............. (-13.54 = -14.23 + 0.70)

| Location | 11,626,946 – 11,627,055 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -19.38 |
| Consensus MFE | -14.95 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
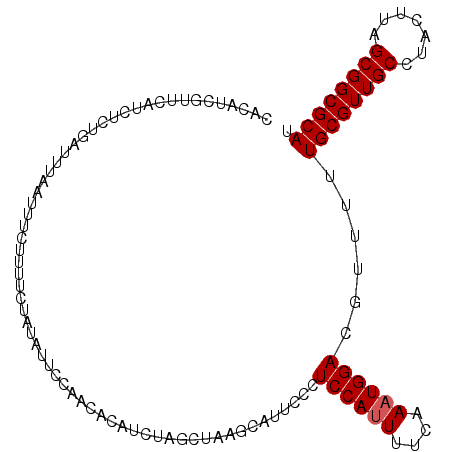
>2L_DroMel_CAF1 11626946 109 - 22407834 CACCUCGUUCAUUUCUGAUUUAAUUUCUUUGCUAUACUUCAACACAUCUAGCUAAGCAUUCCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAU .....((((((((..(((...........((((...((...........))...))))............)))))))))))....((((((((.......)))))))). ( -21.30) >DroSec_CAF1 80298 109 - 1 UACAUCAUUCUUCUCUGAUUUAAUUUCUUUCCUAUAUUCCAACACAUCUAGCUAAGCAUUCCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAU ...((((........))))............................................((((((....))))))......((((((((.......)))))))). ( -15.90) >DroSim_CAF1 84068 109 - 1 CACAUCGUUCAUCUCUGAUUUAAUUUCUUUUCUAUAUUCCAACACAUCUAGCUAAGCAUUGCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAU ..................................................((...(((..((.((((((....)))))).))...)))(((((.......))))))).. ( -18.10) >DroEre_CAF1 110601 109 - 1 CACCUCGUUCAUCUCUGAUUUAAUUUAUUUUCUGUGUUCCAACACAUCCAGCUUAGCAUUCCCUCCAGUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAU .....(((((((..(((...............(((((....)))))....((...))........)))......)))))))....((((((((.......)))))))). ( -22.20) >consensus CACAUCGUUCAUCUCUGAUUUAAUUUCUUUUCUAUAUUCCAACACAUCUAGCUAAGCAUUCCCUCCAUUUUCAAAUGGACGUUUUUGCGUUGCCUACUUAGCGGCGCAU ...............................................................((((((....))))))......((((((((.......)))))))). (-14.95 = -15.20 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:41 2006